Sexual recombination is a signature of a persisting malaria epidemic in Peru
- PMID: 22039962
- PMCID: PMC3231964
- DOI: 10.1186/1475-2875-10-329
Sexual recombination is a signature of a persisting malaria epidemic in Peru
Abstract
Background: The aim of this study was to consider the impact that multi-clone, complex infections have on a parasite population structure in a low transmission setting. In general, complexity of infection (minimum number of clones within an infection) and the overall population level diversity is expected to be minimal in low transmission settings. Additionally, the parasite population structure is predicted to be clonal, rather than sexual due to infrequent parasite inoculation and lack of recombination between genetically distinct clones. However, in this low transmission of the Peruvian Amazon, complex infections are becoming more frequent, in spite of decreasing infection prevalence. In this study, it was hypothesized that sexual recombination between distinct clonal lineages of Plasmodium falciparum parasites were altering the subpopulation structure and effectively maintaining the population-level diversity.
Methods: Fourteen microsatellite markers were chosen to describe the genetic diversity in 313 naturally occurring P. falciparum infections from Peruvian Amazon. The population and subpopulation structure was characterized by measuring: clusteredness, expected heterozygosity (He), allelic richness, private allelic richness, and linkage disequilibrium. Next, microsatellite haplotypes and alleles were correlated with P. falciparum merozoite surface protein 1 Block 2 (Pfmsp1-B2) to examine the presence of recombinant microsatellite haplotypes.
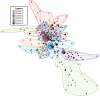
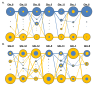
Results: The parasite population structure consists of six genetically diverse subpopulations of clones, called "clusters". Clusters 1, 3, 4, and 6 have unique haplotypes that exceed 70% of the total number of clones within each cluster, while Clusters 2 and 5 have a lower proportion of unique haplotypes, but still exceed 46%. By measuring the He, allelic richness, and private allelic richness within each of the six subpopulations, relatively low levels of genetic diversity within each subpopulation (except Cluster 4) are observed. This indicated that the number of alleles, and not the combination of alleles, are limited. Next, the standard index of association (IAS) was measured, which revealed a significant decay in linkage disequilibrium (LD) associated with Cluster 6, which is indicative of independent assortment of alleles. This decay in LD is a signature of this subpopulation approaching linkage equilibrium by undergoing sexual recombination. To trace possible recombination events, the two most frequent microsatellite haplotypes observed over time (defined by either a K1 or Mad20) were selected as the progenitors and then potential recombinants were identified in within the natural population.
Conclusions: Contrary to conventional low transmission models, this study provides evidence of a parasite population structure that is superficially defined by a clonal backbone. Sexual recombination does occur and even arguably is responsible for maintaining the substructure of this population.
Figures




References
-
- Anderson TJC, Haubold B, Williams JT, Estrada-Franco JG, Richardson L, Mollinedo R, Bockarie M, Mokili J, Mharakurwa S, French N, Whitworth J, Velez ID, Brockman AH, Nosten F, Ferreira MU, Day KP. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol. 2000;17:1467–1482. - PubMed
-
- Barker GC. Microsatellite DNA: a tool for population genetic analysis. Trans R Soc Trop Med Hyg. 2002;96:S21–S24. - PubMed
-
- Havryliuk T, Ferreira MU. A closer look at multiple-clone Plasmodium vivax infections: detection methods, prevalence and consequences. Mem Inst Oswaldo Cruz. 2009;104:67–73. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

