Ubiquitin and ubiquitin-modified proteins activate the Pseudomonas aeruginosa T3SS cytotoxin, ExoU
- PMID: 22040088
- PMCID: PMC3237844
- DOI: 10.1111/j.1365-2958.2011.07904.x
Ubiquitin and ubiquitin-modified proteins activate the Pseudomonas aeruginosa T3SS cytotoxin, ExoU
Abstract
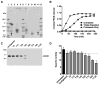
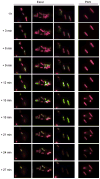
Pseudomonas aeruginosa is an opportunistic Gram-negative pathogen that possesses a type III secretion system (T3SS) critical for evading innate immunity and establishing acute infections in compromised patients. Our research has focused on the structure-activity relationships of ExoU, the most toxic and destructive type III effector produced by P. aeruginosa. ExoU possesses phospholipase activity, which is detectable in vitro only when a eukaryotic cofactor is provided with membrane substrates. We report here that a subpopulation of ubiquitylated yeast SOD1 and other ubiquitylated mammalian proteins activate ExoU. Phospholipase activity was detected using purified ubiquitin of various chain lengths and linkage types; however, free monoubiquitin is sufficient in a genetically engineered dual expression system. The use of ubiquitin by a bacterial enzyme as an activator is unprecedented and represents a new aspect in the manipulation of the eukaryotic ubiquitin system to facilitate bacterial replication and dissemination.
© 2011 Blackwell Publishing Ltd.
Figures

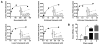
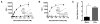


References
-
- Baboshina OV, Haas AL. Novel multiubiquitin chain linkages catalyzed by the conjugating enzymes E2EPF and RAD6 are recognized by 26 S proteasome subunit 5. J Biol Chem. 1996;271:2823–2831. - PubMed
-
- Bohnsack RN, Haas AL. Conservation in the mechanism of Nedd8 activation by the human AppBp1-Uba3 heterodimer. J Biol Chem. 2003;278:26823–26830. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

