MolabIS--an integrated information system for storing and managing molecular genetics data
- PMID: 22040322
- PMCID: PMC3268772
- DOI: 10.1186/1471-2105-12-425
MolabIS--an integrated information system for storing and managing molecular genetics data
Abstract
Background: Long-term sample storage, tracing of data flow and data export for subsequent analyses are of great importance in genetics studies. Therefore, molecular labs do need a proper information system to handle an increasing amount of data from different projects.
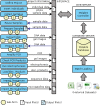
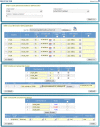
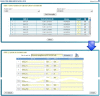
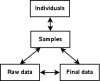
Results: We have developed a molecular labs information management system (MolabIS). It was implemented as a web-based system allowing the users to capture original data at each step of their workflow. MolabIS provides essential functionality for managing information on individuals, tracking samples and storage locations, capturing raw files, importing final data from external files, searching results, accessing and modifying data. Further important features are options to generate ready-to-print reports and convert sequence and microsatellite data into various data formats, which can be used as input files in subsequent analyses. Moreover, MolabIS also provides a tool for data migration.
Conclusions: MolabIS is designed for small-to-medium sized labs conducting Sanger sequencing and microsatellite genotyping to store and efficiently handle a relative large amount of data. MolabIS not only helps to avoid time consuming tasks but also ensures the availability of data for further analyses. The software is packaged as a virtual appliance which can run on different platforms (e.g. Linux, Windows). MolabIS can be distributed to a wide range of molecular genetics labs since it was developed according to a general data model. Released under GPL, MolabIS is freely available at http://www.molabis.org.
Figures

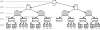



Similar articles
-
Laboratory Information Management Software for genotyping workflows: applications in high throughput crop genotyping.BMC Bioinformatics. 2006 Aug 17;7:383. doi: 10.1186/1471-2105-7-383. BMC Bioinformatics. 2006. PMID: 16914063 Free PMC article.
-
DraGnET: software for storing, managing and analyzing annotated draft genome sequence data.BMC Bioinformatics. 2010 Feb 22;11:100. doi: 10.1186/1471-2105-11-100. BMC Bioinformatics. 2010. PMID: 20175920 Free PMC article.
-
QDD: a user-friendly program to select microsatellite markers and design primers from large sequencing projects.Bioinformatics. 2010 Feb 1;26(3):403-4. doi: 10.1093/bioinformatics/btp670. Epub 2009 Dec 10. Bioinformatics. 2010. PMID: 20007741
-
MetaLIMS, a simple open-source laboratory information management system for small metagenomic labs.Gigascience. 2017 Jun 1;6(6):1-6. doi: 10.1093/gigascience/gix025. Gigascience. 2017. PMID: 28430964 Free PMC article.
-
An information-driven approach to pharmacogenomics.Pharmacogenomics. 2005 Jul;6(5):473-80. doi: 10.2217/14622416.6.5.473. Pharmacogenomics. 2005. PMID: 16013997 Review.
Cited by
-
COVID-19 pandemic: Is it the right time to develop interconnected national biomedical registries?Genomics Inform. 2021 Dec;19(4):e50. doi: 10.5808/gi.21021. Epub 2021 Dec 31. Genomics Inform. 2021. PMID: 35012292 Free PMC article. No abstract available.
-
SNPflow: a lightweight application for the processing, storing and automatic quality checking of genotyping assays.PLoS One. 2013;8(3):e59508. doi: 10.1371/journal.pone.0059508. Epub 2013 Mar 19. PLoS One. 2013. PMID: 23527209 Free PMC article.
-
Efficient sample tracking with OpenLabFramework.Sci Rep. 2014 Mar 4;4:4278. doi: 10.1038/srep04278. Sci Rep. 2014. PMID: 24589879 Free PMC article.
-
QTREDS: a Ruby on Rails-based platform for omics laboratories.BMC Bioinformatics. 2014;15 Suppl 1(Suppl 1):S13. doi: 10.1186/1471-2105-15-S1-S13. Epub 2014 Jan 10. BMC Bioinformatics. 2014. PMID: 24564791 Free PMC article.
-
Phytotracker, an information management system for easy recording and tracking of plants, seeds and plasmids.Plant Methods. 2012 Oct 13;8(1):43. doi: 10.1186/1746-4811-8-43. Plant Methods. 2012. PMID: 23062011 Free PMC article.
References
-
- Baumung R, Simianer H, Hoffmann I. Genetic diversity studies in farm animals - a survey. J of Anim Breed and Genet. 2004;121(6):361–373. doi: 10.1111/j.1439-0388.2004.00479.x. - DOI
-
- Granevitze Z, Hillel J, Chen GH, Cuc NTK, Feldman M, Eding H, Weigend S. Genetic diversity within chicken populations from different continents and management histories. Animal Genetics. 2007;36(6):576–583. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

