Cell-autonomous circadian clock of hepatocytes drives rhythms in transcription and polyamine synthesis
- PMID: 22042857
- PMCID: PMC3215069
- DOI: 10.1073/pnas.1115753108
Cell-autonomous circadian clock of hepatocytes drives rhythms in transcription and polyamine synthesis
Abstract
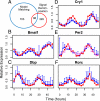
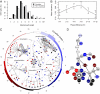
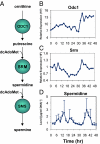
The circadian clock generates daily rhythms in mammalian liver processes, such as glucose and lipid homeostasis, xenobiotic metabolism, and regeneration. The mechanisms governing these rhythms are not well understood, particularly the distinct contributions of the cell-autonomous clock and central pacemaker to rhythmic liver physiology. Through microarray expression profiling in Met murine hepatocytes (MMH)-D3, we identified over 1,000 transcripts that exhibit circadian oscillations, demonstrating that the cell-autonomous clock can drive many rhythms, and that MMH-D3 is a valid circadian model system. The genes represented by these circadian transcripts displayed both cophasic and antiphasic organization within a protein-protein interaction network, suggesting the existence of competition for binding sites or partners by genes of disparate transcriptional phases. Multiple pathways displayed enrichment in MMH-D3 circadian transcripts, including the polyamine synthesis module of the glutathione metabolic pathway. The polyamine synthesis module, which is highly associated with cell proliferation and whose products are required for initiation of liver regeneration, includes enzymes whose transcripts exhibit circadian oscillations, such as ornithine decarboxylase and spermidine synthase. Metabolic profiling revealed that the enzymatic product of spermidine synthase, spermidine, cycles as well. Thus, the cell-autonomous hepatocyte clock can drive a significant amount of transcriptional rhythms and orchestrate physiologically relevant modules such as polyamine synthesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Hastings MH, Reddy AB, Maywood ES. A clockwork web: Circadian timing in brain and periphery, in health and disease. Nat Rev Neurosci. 2003;4:649–661. - PubMed
-
- Zhang EE, Kay SA. Clocks not winding down: Unravelling circadian networks. Nat Rev Mol Cell Biol. 2010;11:764–776. - PubMed
-
- Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

