A systematic screen for micro-RNAs regulating the canonical Wnt pathway
- PMID: 22043311
- PMCID: PMC3197157
- DOI: 10.1371/journal.pone.0026257
A systematic screen for micro-RNAs regulating the canonical Wnt pathway
Abstract
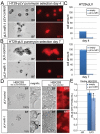
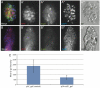
MicroRNAs (miRs) and the canonical Wnt pathway are known to be dysregulated in human cancers and play key roles during cancer initiation and progression. To identify miRs that can modulate the activity of the Wnt pathway we performed a cell-based overexpression screen of 470 miRs in human HEK293 cells. We identified 38 candidate miRs that either activate or repress the canonical Wnt pathway. A literature survey of all verified candidate miRs revealed that the Wnt-repressing miRs tend to be anti-oncomiRs and down-regulated in cancers while Wnt-activating miRs tend to be oncomiRs and upregulated during tumorigenesis. Epistasis-based functional validation of three candidate miRs, miR-1, miR-25 and miR-613, confirmed their inhibitory role in repressing the Wnt pathway and suggest that while miR-25 may function at the level of â-catenin (β-cat), miR-1 and miR-613 act upstream of β-cat. Both miR-25 and miR-1 inhibit cell proliferation and viability during selection of human colon cancer cell lines that exhibit dysregulated Wnt signaling. Finally, transduction of miR-1 expressing lentiviruses into primary mammary organoids derived from Conductin-lacZ mice significantly reduced the expression of the Wnt-sensitive β-gal reporter. In summary, these findings suggest the potential use of Wnt-modulating miRs as diagnostic and therapeutic tools in Wnt-dependent diseases, such as cancer.
Conflict of interest statement
Figures



References
-
- Moser AR, Luongo C, Gould KA, McNeley MK, Shoemaker AR, et al. ApcMin: a mouse model for intestinal and mammary tumorigenesis. Eur J Cancer. 1995;31A:1061–1064. - PubMed
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Liu Q, Paroo Z. Biochemical Principles of Small RNA Pathways. Annu Rev Biochem 2010 - PubMed
-
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

