Minimal functional sites allow a classification of zinc sites in proteins
- PMID: 22043316
- PMCID: PMC3197139
- DOI: 10.1371/journal.pone.0026325
Minimal functional sites allow a classification of zinc sites in proteins
Abstract
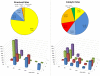
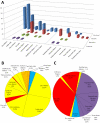
Zinc is indispensable to all forms of life as it is an essential component of many different proteins involved in a wide range of biological processes. Not differently from other metals, zinc in proteins can play different roles that depend on the features of the metal-binding site. In this work, we describe zinc sites in proteins with known structure by means of three-dimensional templates that can be automatically extracted from PDB files and consist of the protein structure around the metal, including the zinc ligands and the residues in close spatial proximity to the ligands. This definition is devised to intrinsically capture the features of the local protein environment that can affect metal function, and corresponds to what we call a minimal functional site (MFS). We used MFSs to classify all zinc sites whose structures are available in the PDB and combined this classification with functional annotation as available in the literature. We classified 77% of zinc sites into ten clusters, each grouping zinc sites with structures that are highly similar, and an additional 16% into seven pseudo-clusters, each grouping zinc sites with structures that are only broadly similar. Sites where zinc plays a structural role are predominant in eight clusters and in two pseudo-clusters, while sites where zinc plays a catalytic role are predominant in two clusters and in five pseudo-clusters. We also analyzed the amino acid composition of the coordination sphere of zinc as a function of its role in the protein, highlighting trends and exceptions. In a period when the number of known zinc proteins is expected to grow further with the increasing awareness of the cellular mechanisms of zinc homeostasis, this classification represents a valuable basis for structure-function studies of zinc proteins, with broad applications in biochemistry, molecular pharmacology and de novo protein design.
Conflict of interest statement
Figures




References
-
- Bertini I, Sigel A, Sigel H. Handbook on Metalloproteins. New York: Marcel Dekker; 2001.
-
- Frausto da Silva JJR, Williams RJP. The biological chemistry of the elements: the inorganic chemistry of life. New York: Oxford University Press; 2001.
-
- Andreini C, Banci L, Bertini I, Rosato A. Counting the zinc proteins encoded in the human genome. J Proteome Res. 2006;5:196–201. - PubMed
-
- Andreini C, Banci L, Bertini I, Rosato A. Zinc through the three domains of life. J Proteome Res. 2006;5:3173–3178. - PubMed
-
- Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM. Metal ions in biological catalysis: from enzyme databases to general principles. J Biol Inorg Chem. 2008;13:1205–1218. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

