Projection to latent pathways (PLP): a constrained projection to latent variables (PLS) method for elementary flux modes discrimination
- PMID: 22044634
- PMCID: PMC3750108
- DOI: 10.1186/1752-0509-5-181
Projection to latent pathways (PLP): a constrained projection to latent variables (PLS) method for elementary flux modes discrimination
Abstract
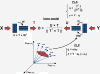
Background: Elementary flux modes (EFM) are unique and non-decomposable sets of metabolic reactions able to operate coherently in steady-state. A metabolic network has in general a very high number of EFM reflecting the typical functional redundancy of biological systems. However, most of these EFM are either thermodynamically unfeasible or inactive at pre-set environmental conditions.
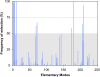
Results: Here we present a new algorithm that discriminates the "active" set of EFM on the basis of dynamic envirome data. The algorithm merges together two well-known methods: projection to latent structures (PLS) and EFM analysis, and is therefore termed projection to latent pathways (PLP). PLP has two concomitant goals: (1) maximisation of correlation between EFM weighting factors and measured envirome data and (2) minimisation of redundancy by eliminating EFM with low correlation with the envirome.
Conclusions: Overall, our results demonstrate that PLP slightly outperforms PLS in terms of predictive power. But more importantly, PLP is able to discriminate the subset of EFM with highest correlation with the envirome, thus providing in-depth knowledge of how the environment controls core cellular functions. This offers a significant advantage over PLS since its abstract structure cannot be associated with the underlying biological structure.
Figures






References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

