The pea TCP transcription factor PsBRC1 acts downstream of Strigolactones to control shoot branching
- PMID: 22045922
- PMCID: PMC3252107
- DOI: 10.1104/pp.111.182725
The pea TCP transcription factor PsBRC1 acts downstream of Strigolactones to control shoot branching
Abstract
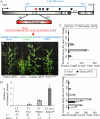
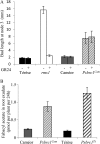
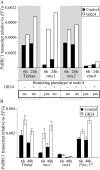
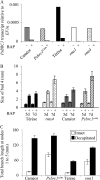
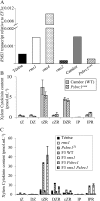
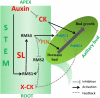
The function of PsBRC1, the pea (Pisum sativum) homolog of the maize (Zea mays) TEOSINTE BRANCHED1 and the Arabidopsis (Arabidopsis thaliana) BRANCHED1 (AtBRC1) genes, was investigated. The pea Psbrc1 mutant displays an increased shoot-branching phenotype, is able to synthesize strigolactone (SL), and does not respond to SL application. The level of pleiotropy of the SL-deficient ramosus1 (rms1) mutant is higher than in the Psbrc1 mutant, rms1 exhibiting a relatively dwarf phenotype and more extensive branching at upper nodes. The PsBRC1 gene is mostly expressed in the axillary bud and is transcriptionally up-regulated by direct application of the synthetic SL GR24 and down-regulated by the cytokinin (CK) 6-benzylaminopurine. The results suggest that PsBRC1 may have a role in integrating SL and CK signals and that SLs act directly within the bud to regulate its outgrowth. However, the Psbrc1 mutant responds to 6-benzylaminopurine application and decapitation by increasing axillary bud length, implicating a PsBRC1-independent component of the CK response in sustained bud growth. In contrast to other SL-related mutants, the Psbrc1 mutation does not cause a decrease in the CK zeatin riboside in the xylem sap or a strong increase in RMS1 transcript levels, suggesting that the RMS2-dependent feedback is not activated in this mutant. Surprisingly, the double rms1 Psbrc1 mutant displays a strong increase in numbers of branches at cotyledonary nodes, whereas branching at upper nodes is not significantly higher than the branching in rms1. This phenotype indicates a localized regulation of branching at these nodes specific to pea.
Figures
References
-
- Aggarwal P, Padmanabhan B, Bhat A, Sarvepalli K, Sadhale PP, Nath U. (2011) The TCP4 transcription factor of Arabidopsis blocks cell division in yeast at G1→S transition. Biochem Biophys Res Commun 410: 276–281 - PubMed
-
- Arite T, Iwata H, Ohshima K, Maekawa M, Nakajima M, Kojima M, Sakakibara H, Kyozuka J. (2007) DWARF10, an RMS1/MAX4/DAD1 ortholog, controls lateral bud outgrowth in rice. Plant J 51: 1019–1029 - PubMed
-
- Arumingtyas EL, Floyd RS, Gregory MJ, Murfet IC. (1992) Branching in Pisum: inheritance and allelism tests with 17 ramosus mutants. Pisum Genet 24: 17–31
-
- Bangerth F. (1994) Response of cytokinin concentration in the xylem exudate of bean (Phaseolus vulgaris L.) plants to decapitation and auxin treatment, and relationship to apical dominance. Planta 194: 439–442
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

