Coordinated destruction of cellular messages in translation complexes by the gammaherpesvirus host shutoff factor and the mammalian exonuclease Xrn1
- PMID: 22046136
- PMCID: PMC3203186
- DOI: 10.1371/journal.ppat.1002339
Coordinated destruction of cellular messages in translation complexes by the gammaherpesvirus host shutoff factor and the mammalian exonuclease Xrn1
Abstract
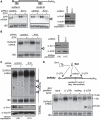
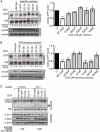
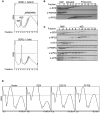
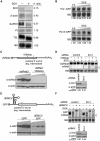
Several viruses encode factors that promote host mRNA degradation to silence gene expression. It is unclear, however, whether cellular mRNA turnover pathways are engaged to assist in this process. In Kaposi's sarcoma-associated herpesvirus this phenotype is enacted by the host shutoff factor SOX. Here we show that SOX-induced mRNA turnover is a two-step process, in which mRNAs are first cleaved internally by SOX itself then degraded by the cellular exonuclease Xrn1. SOX therefore bypasses the regulatory steps of deadenylation and decapping normally required for Xrn1 activation. SOX is likely recruited to translating mRNAs, as it cosediments with translation initiation complexes and depletes polysomes. Cleaved mRNA intermediates accumulate in the 40S fraction, indicating that recognition occurs at an early stage of translation. This is the first example of a viral protein commandeering cellular mRNA turnover pathways to destroy host mRNAs, and suggests that Xrn1 is poised to deplete messages undergoing translation in mammalian cells.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

