Comparative genomic analysis of human fungal pathogens causing paracoccidioidomycosis
- PMID: 22046142
- PMCID: PMC3203195
- DOI: 10.1371/journal.pgen.1002345
Comparative genomic analysis of human fungal pathogens causing paracoccidioidomycosis
Abstract
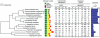
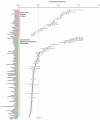
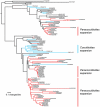
Paracoccidioides is a fungal pathogen and the cause of paracoccidioidomycosis, a health-threatening human systemic mycosis endemic to Latin America. Infection by Paracoccidioides, a dimorphic fungus in the order Onygenales, is coupled with a thermally regulated transition from a soil-dwelling filamentous form to a yeast-like pathogenic form. To better understand the genetic basis of growth and pathogenicity in Paracoccidioides, we sequenced the genomes of two strains of Paracoccidioides brasiliensis (Pb03 and Pb18) and one strain of Paracoccidioides lutzii (Pb01). These genomes range in size from 29.1 Mb to 32.9 Mb and encode 7,610 to 8,130 genes. To enable genetic studies, we mapped 94% of the P. brasiliensis Pb18 assembly onto five chromosomes. We characterized gene family content across Onygenales and related fungi, and within Paracoccidioides we found expansions of the fungal-specific kinase family FunK1. Additionally, the Onygenales have lost many genes involved in carbohydrate metabolism and fewer genes involved in protein metabolism, resulting in a higher ratio of proteases to carbohydrate active enzymes in the Onygenales than their relatives. To determine if gene content correlated with growth on different substrates, we screened the non-pathogenic onygenale Uncinocarpus reesii, which has orthologs for 91% of Paracoccidioides metabolic genes, for growth on 190 carbon sources. U. reesii showed growth on a limited range of carbohydrates, primarily basic plant sugars and cell wall components; this suggests that Onygenales, including dimorphic fungi, can degrade cellulosic plant material in the soil. In addition, U. reesii grew on gelatin and a wide range of dipeptides and amino acids, indicating a preference for proteinaceous growth substrates over carbohydrates, which may enable these fungi to also degrade animal biomass. These capabilities for degrading plant and animal substrates suggest a duality in lifestyle that could enable pathogenic species of Onygenales to transfer from soil to animal hosts.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Martinez R. Paracoccidioidomycosis: the dimension of the problem of a neglected disease. Rev Soc Bras Med Trop. 2010;43:480. - PubMed
-
- Wanke B, Londero AT. Epidemiology and paracoccidioidomycosis infection. In: Franco M, Lacaz CdS, Restrepo-Moreno A, Del Negro G, editors. Paracoccidioidomycosis. Boca Raton: CRC Press; 1994. pp. 109–120.
-
- Hotez PJ, Bottazzi ME, Franco-Paredes C, Ault SK, Periago MR. The neglected tropical diseases of Latin America and the Caribbean: a review of disease burden and distribution and a roadmap for control and elimination. PLoS Negl Trop Dis. 2008;2:e300. doi: 10.1371/journal.pntd.0000300. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

