Oscillatory dynamics of cell cycle proteins in single yeast cells analyzed by imaging cytometry
- PMID: 22046265
- PMCID: PMC3202528
- DOI: 10.1371/journal.pone.0026272
Oscillatory dynamics of cell cycle proteins in single yeast cells analyzed by imaging cytometry
Abstract
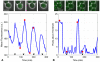
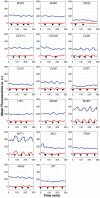
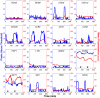
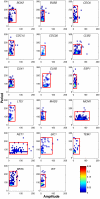
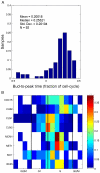
Progression through the cell division cycle is orchestrated by a complex network of interacting genes and proteins. Some of these proteins are known to fluctuate periodically during the cell cycle, but a systematic study of the fluctuations of a broad sample of cell-cycle proteins has not been made until now. Using time-lapse fluorescence microscopy, we profiled 16 strains of budding yeast, each containing GFP fused to a single gene involved in cell cycle regulation. The dynamics of protein abundance and localization were characterized by extracting the amplitude, period, and other indicators from a series of images. Oscillations of protein abundance could clearly be identified for Cdc15, Clb2, Cln1, Cln2, Mcm1, Net1, Sic1, and Whi5. The period of oscillation of the fluorescently tagged proteins is generally in good agreement with the inter-bud time. The very strong oscillations of Net1 and Mcm1 expression are remarkable since little is known about the temporal expression of these genes. By collecting data from large samples of single cells, we quantified some aspects of cell-to-cell variability due presumably to intrinsic and extrinsic noise affecting the cell cycle.
Conflict of interest statement
Figures
References
-
- Morgan DO. The cell cycle : principles of control. London: New Science Press, Ltd; 2007.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

