A novel evolution-based method for detecting gene-gene interactions
- PMID: 22046286
- PMCID: PMC3201950
- DOI: 10.1371/journal.pone.0026435
A novel evolution-based method for detecting gene-gene interactions
Abstract
Background: The rapid advance in large-scale SNP-chip technologies offers us great opportunities in elucidating the genetic basis of complex diseases. Methods for large-scale interactions analysis have been under development from several sources. Due to several difficult issues (e.g., sparseness of data in high dimensions and low replication or validation rate), development of fast, powerful and robust methods for detecting various forms of gene-gene interactions continues to be a challenging task.
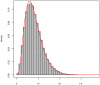
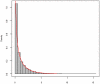
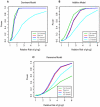
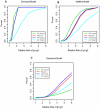
Methodology/principal findings: In this article, we have developed an evolution-based method to search for genome-wide epistasis in a case-control design. From an evolutionary perspective, we view that human diseases originate from ancient mutations and consider that the underlying genetic variants play a role in differentiating human population into the healthy and the diseased. Based on this concept, traditional evolutionary measure, fixation index (Fst) for two unlinked loci, which measures the genetic distance between populations, should be able to reveal the responsible genetic interplays for disease traits. To validate our proposal, we first investigated the theoretical distribution of Fst by using extensive simulations. Then, we explored its power for detecting gene-gene interactions via SNP markers, and compared it with the conventional Pearson Chi-square test, mutual information based test and linkage disequilibrium based test under several disease models. The proposed evolution-based method outperformed these compared methods in dominant and additive models, no matter what the disease allele frequencies were. However, its performance was relatively poor in a recessive model. Finally, we applied the proposed evolution-based method to analysis of a published dataset. Our results showed that the P value of the Fst -based statistic is smaller than those obtained by the LD-based statistic or Poisson regression models.
Conclusions/significance: With rapidly growing large-scale genetic association studies, the proposed evolution-based method can be a promising tool in the identification of epistatic effects.
Conflict of interest statement
Figures


References
-
- Marchini J, Donnelly P, Cardon LR. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat Genet. 2005;37:413–417. - PubMed
-
- Hoh J, Ott J. Mathematical multi-locus approaches to localizing complex human trait genes. Nat Rev Genet. 2003;4:701–709. - PubMed
-
- Camp NJ, Slattery ML. Classification tree analysis: a statistical tool to investigate risk factor interactions with an example for colon cancer (United States). Cancer Causes Control. 2002;13:813–823. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

