Exploring the diversity of Gardnerella vaginalis in the genitourinary tract microbiota of monogamous couples through subtle nucleotide variation
- PMID: 22046340
- PMCID: PMC3201972
- DOI: 10.1371/journal.pone.0026732
Exploring the diversity of Gardnerella vaginalis in the genitourinary tract microbiota of monogamous couples through subtle nucleotide variation
Abstract
Background: Bacterial vaginosis (BV) is an enigmatic disease of unknown origin that affects a large percentage of women. The vaginal microbiota of women with BV is associated with serious sequelae, including abnormal pregnancies. The etiology of BV is not fully understood, however, it has been suggested that it is transmissible, and that G. vaginalis may be an etiological agent. Studies using enzymatic assays to define G. vaginalis biotypes, as well as more recent genomic comparisons of G. vaginalis isolates from symptomatic and asymptomatic women, suggest that particular G. vaginalis strains may play a key role in the pathogenesis of BV.
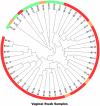
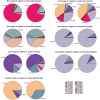
Methodology/principal findings: To explore G. vaginalis diversity, distribution and sexual transmission, we developed a Shannon entropy-based method to analyze low-level sequence variation in 65,710 G. vaginalis 16S rRNA gene segments that were PCR-amplified from vaginal samples of 53 monogamous women and from urethral and penile skin samples of their male partners. We observed a high degree of low-level diversity among G. vaginalis sequences with a total of 46 unique sequence variants (oligotypes), and also found strong correlations of these oligotypes between sexual partners. Even though Gram stain-defined normal and some Gram stain-defined intermediate oligotype profiles clustered together in UniFrac analysis, no single G. vaginalis oligotype was found to be specific to BV or normal vaginal samples.
Conclusions: This study describes a novel method for investigating G. vaginalis diversity at a low level of taxonomic discrimination. The findings support cultivation-based studies that indicate sexual partners harbor the same strains of G. vaginalis. This study also highlights the fact that a few, reproducible nucleotide variations within the 16S rRNA gene can reveal clinical or epidemiological associations that would be missed by genus-level or species-level categorization of 16S rRNA data.
Conflict of interest statement
Figures

Similar articles
-
Cloning of 16S rRNA genes amplified from normal and disturbed vaginal microflora suggests a strong association between Atopobium vaginae, Gardnerella vaginalis and bacterial vaginosis.BMC Microbiol. 2004 Apr 21;4:16. doi: 10.1186/1471-2180-4-16. BMC Microbiol. 2004. PMID: 15102329 Free PMC article.
-
Resolution and characterization of distinct cpn60-based subgroups of Gardnerella vaginalis in the vaginal microbiota.PLoS One. 2012;7(8):e43009. doi: 10.1371/journal.pone.0043009. Epub 2012 Aug 10. PLoS One. 2012. PMID: 22900080 Free PMC article.
-
Bacterial communities in penile skin, male urethra, and vaginas of heterosexual couples with and without bacterial vaginosis.Microbiome. 2016 Apr 19;4:16. doi: 10.1186/s40168-016-0161-6. Microbiome. 2016. PMID: 27090518 Free PMC article.
-
Genetic Heterogeneity and Taxonomic Diversity among Gardnerella Species.Trends Microbiol. 2020 Mar;28(3):202-211. doi: 10.1016/j.tim.2019.10.002. Epub 2019 Nov 4. Trends Microbiol. 2020. PMID: 31699644 Review.
-
Research Progress on the Correlation Between Gardnerella Typing and Bacterial Vaginosis.Front Cell Infect Microbiol. 2022 Mar 25;12:858155. doi: 10.3389/fcimb.2022.858155. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35402309 Free PMC article. Review.
Cited by
-
The Relevance of Host Gut Microbiome Signature Alterations on de novo Fatty Acids Synthesis in Patients with Multi-Drug Resistant Tuberculosis.Infect Drug Resist. 2022 Sep 21;15:5589-5600. doi: 10.2147/IDR.S372122. eCollection 2022. Infect Drug Resist. 2022. PMID: 36168638 Free PMC article.
-
Changes in gut microbiota in rats fed a high fat diet correlate with obesity-associated metabolic parameters.PLoS One. 2015 May 18;10(5):e0126931. doi: 10.1371/journal.pone.0126931. eCollection 2015. PLoS One. 2015. PMID: 25992554 Free PMC article.
-
Distinct systemic microbiome and microbial translocation are associated with plasma level of anti-CD4 autoantibody in HIV infection.Sci Rep. 2018 Aug 27;8(1):12863. doi: 10.1038/s41598-018-31116-y. Sci Rep. 2018. PMID: 30150778 Free PMC article.
-
Sex differences in the fecal microbiome and hippocampal glial morphology following diet and antibiotic treatment.PLoS One. 2022 Apr 6;17(4):e0265850. doi: 10.1371/journal.pone.0265850. eCollection 2022. PLoS One. 2022. PMID: 35385494 Free PMC article.
-
Spoilage Investigation of Chill Stored Meagre (Argyrosomus regius) Using Modern Microbiological and Analytical Techniques.Foods. 2021 Dec 15;10(12):3109. doi: 10.3390/foods10123109. Foods. 2021. PMID: 34945660 Free PMC article.
References
-
- Frank DN, Pace NR. Gastrointestinal microbiology enters the metagenomics era. Curr Opin Gastroenterol. 2008;24:4–10. - PubMed
-
- Savage DC. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. - PubMed
-
- Berg RD. The indigenous gastrointestinal microflora. Trends Microbiol. 1996;4:430–435. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

