Predicting residue-residue contacts and helix-helix interactions in transmembrane proteins using an integrative feature-based random forest approach
- PMID: 22046350
- PMCID: PMC3203928
- DOI: 10.1371/journal.pone.0026767
Predicting residue-residue contacts and helix-helix interactions in transmembrane proteins using an integrative feature-based random forest approach
Abstract
Integral membrane proteins constitute 25-30% of genomes and play crucial roles in many biological processes. However, less than 1% of membrane protein structures are in the Protein Data Bank. In this context, it is important to develop reliable computational methods for predicting the structures of membrane proteins. Here, we present the first application of random forest (RF) for residue-residue contact prediction in transmembrane proteins, which we term as TMhhcp. Rigorous cross-validation tests indicate that the built RF models provide a more favorable prediction performance compared with two state-of-the-art methods, i.e., TMHcon and MEMPACK. Using a strict leave-one-protein-out jackknifing procedure, they were capable of reaching the top L/5 prediction accuracies of 49.5% and 48.8% for two different residue contact definitions, respectively. The predicted residue contacts were further employed to predict interacting helical pairs and achieved the Matthew's correlation coefficients of 0.430 and 0.424, according to two different residue contact definitions, respectively. To facilitate the academic community, the TMhhcp server has been made freely accessible at http://protein.cau.edu.cn/tmhhcp.
Conflict of interest statement
Figures
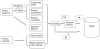


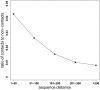

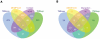

References
-
- Yildirim MA, Goh KI, Cusick ME, Barabasi AL, Vidal M. Drug-target network. Nature Biotechnology. 2007;25:1119–1126. - PubMed
-
- Doerr A. Membrane protein structures. Nature Methods. 2009;6:35–35.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

