Rates of mutation and host transmission for an Escherichia coli clone over 3 years
- PMID: 22046404
- PMCID: PMC3203180
- DOI: 10.1371/journal.pone.0026907
Rates of mutation and host transmission for an Escherichia coli clone over 3 years
Abstract
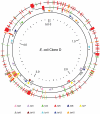
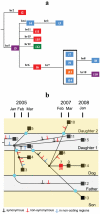
Although over 50 complete Escherichia coli/Shigella genome sequences are available, it is only for closely related strains, for example the O55:H7 and O157:H7 clones of E. coli, that we can assign differences to individual evolutionary events along specific lineages. Here we sequence the genomes of 14 isolates of a uropathogenic E. coli clone that persisted for 3 years within a household, including a dog, causing a urinary tract infection (UTI) in the dog after 2 years. The 20 mutations observed fit a single tree that allows us to estimate the mutation rate to be about 1.1 per genome per year, with minimal evidence for adaptive change, including in relation to the UTI episode. The host data also imply at least 6 host transfer events over the 3 years, with 2 lineages present over much of that period. To our knowledge, these are the first direct measurements for a clone in a well-defined host community that includes rates of mutation and host transmission. There is a concentration of non-synonymous mutations associated with 2 transfers to the dog, suggesting some selection pressure from the change of host. However, there are no changes to which we can attribute the UTI event in the dog, which suggests that this occurrence after 2 years of the clone being in the household may have been due to chance, or some unknown change in the host or environment. The ability of a UTI strain to persist for 2 years and also to transfer readily within a household has implications for epidemiology, diagnosis, and clinical intervention.
Conflict of interest statement
Figures

References
-
- Johnson JR, Clabots C. Sharing of virulent Escherichia coli clones among household members of a woman with acute cystitis. Clin Infect Dis. 2006;43:e101–108. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

