Structural and evolutionary divergence of eukaryotic protein kinases in Apicomplexa
- PMID: 22047078
- PMCID: PMC3239843
- DOI: 10.1186/1471-2148-11-321
Structural and evolutionary divergence of eukaryotic protein kinases in Apicomplexa
Abstract
Background: The Apicomplexa constitute an evolutionarily divergent phylum of protozoan pathogens responsible for widespread parasitic diseases such as malaria and toxoplasmosis. Many cellular functions in these medically important organisms are controlled by protein kinases, which have emerged as promising drug targets for parasitic diseases. However, an incomplete understanding of how apicomplexan kinases structurally and mechanistically differ from their host counterparts has hindered drug development efforts to target parasite kinases.
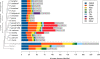
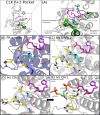
Results: We used the wealth of sequence data recently made available for 15 apicomplexan species to identify the kinome of each species and quantify the evolutionary constraints imposed on each family of apicomplexan kinases. Our analysis revealed lineage-specific adaptations in selected families, namely cyclin-dependent kinase (CDK), calcium-dependent protein kinase (CDPK) and CLK/LAMMER, which have been identified as important in the pathogenesis of these organisms. Bayesian analysis of selective constraints imposed on these families identified the sequence and structural features that most distinguish apicomplexan protein kinases from their homologs in model organisms and other eukaryotes. In particular, in a subfamily of CDKs orthologous to Plasmodium falciparum crk-5, the activation loop contains a novel PTxC motif which is absent from all CDKs outside Apicomplexa. Our analysis also suggests a convergent mode of regulation in a subset of apicomplexan CDPKs and mammalian MAPKs involving a commonly conserved arginine in the αC helix. In all recognized apicomplexan CLKs, we find a set of co-conserved residues involved in substrate recognition and docking that are distinct from metazoan CLKs.
Conclusions: We pinpoint key conserved residues that can be predicted to mediate functional differences from eukaryotic homologs in three identified kinase families. We discuss the structural, functional and evolutionary implications of these lineage-specific variations and propose specific hypotheses for experimental investigation. The apicomplexan-specific kinase features reported in this study can be used in the design of selective kinase inhibitors.
Figures

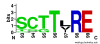

References
-
- Roos DS. Genetics. Themes and variations in apicomplexan parasite biology. Science. 2005;309(5731):72–3. - PubMed
-
- Renslo AR, McKerrow JH. Drug discovery and development for neglected parasitic diseases. Nature chemical biology. 2006;2(12):701–10. - PubMed
-
- Sibley LD. Intracellular parasite invasion strategies. Science. 2004;304(5668):248–53. - PubMed
-
- Hammarton T. The cell cycle of parasitic protozoa: potential for chemotherapeutic exploitation. Progress In Cell Cycle Research. 2003;5:91–101. - PubMed
-
- Doerig C, Abdi A, Bland N, Eschenlauer S, Dorin-Semblat D, Fennell C, Halbert J, Holland Z, Nivez MP, Semblat JP, Sicard A, Reininger L. Malaria: targeting parasite and host cell kinomes. Biochimica et biophysica acta. 2010;1804(3):604–12. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

