Investigation by site-directed mutagenesis of the role of cytochrome P450 2B4 non-active-site residues in protein-ligand interactions based on crystal structures of the ligand-bound enzyme
- PMID: 22051155
- PMCID: PMC3911823
- DOI: 10.1111/j.1742-4658.2011.08411.x
Investigation by site-directed mutagenesis of the role of cytochrome P450 2B4 non-active-site residues in protein-ligand interactions based on crystal structures of the ligand-bound enzyme
Abstract
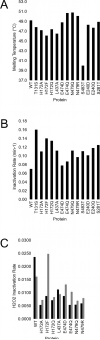
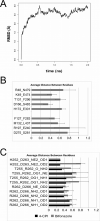
Residues located outside the active site of cytochromes P450 2B have exhibited importance in ligand binding, structural stability and drug metabolism. However, contributions of non-active-site residues to the plasticity of these enzymes are not known. Thus, a systematic investigation was undertaken of unique residue-residue interactions found in crystal structures of P450 2B4 in complex with 4-(4-chlorophenyl)imidazole (4-CPI), a closed conformation, or in complex with bifonazole, an expanded conformation. Nineteen mutants distributed over 11 sites were constructed, expressed in Escherichia coli and purified. Most mutants showed significantly decreased expression, especially in the case of interactions found in the 4-CPI structure. Six mutants (H172A, H172F, H172Q, L437A, E474D and E474Q) were chosen for detailed functional analysis. Among these, the K(s) of H172F for bifonazole was ∼ 20 times higher than for wild-type 2B4, and the K(s) of L437A for 4-CPI was ∼ 50 times higher than for wild-type, leading to significantly altered inhibitor selectivity. Enzyme function was tested with the substrates 7-ethoxy-4-(trifluoromethyl)coumarin, 7-methoxy-4-(trifluoromethyl)coumarin and 7-benzyloxyresorufin (7-BR). H172F was inactive with all three substrates, and L437A did not turn over 7-BR. Furthermore, H172A, H172Q, E474D and E474Q showed large changes in k(cat)/K(M) for each of the three substrates, in some cases up to 50-fold. Concurrent molecular dynamics simulations yielded distances between some of the residues in these putative interaction pairs that are not consistent with contact. The results indicate that small changes in the protein scaffold lead to large differences in solution behavior and enzyme function.
© 2011 The Authors Journal compilation © 2011 FEBS.
Figures



Similar articles
-
Structure of microsomal cytochrome P450 2B4 complexed with the antifungal drug bifonazole: insight into P450 conformational plasticity and membrane interaction.J Biol Chem. 2006 Mar 3;281(9):5973-81. doi: 10.1074/jbc.M511464200. Epub 2005 Dec 21. J Biol Chem. 2006. PMID: 16373351
-
Investigation of the role of cytochrome P450 2B4 active site residues in substrate metabolism based on crystal structures of the ligand-bound enzyme.Arch Biochem Biophys. 2006 Nov 1;455(1):61-7. doi: 10.1016/j.abb.2006.08.024. Epub 2006 Sep 25. Arch Biochem Biophys. 2006. PMID: 17027909 Free PMC article.
-
Structure of mammalian cytochrome P450 2B4 complexed with 4-(4-chlorophenyl)imidazole at 1.9-A resolution: insight into the range of P450 conformations and the coordination of redox partner binding.J Biol Chem. 2004 Jun 25;279(26):27294-301. doi: 10.1074/jbc.M403349200. Epub 2004 Apr 20. J Biol Chem. 2004. PMID: 15100217
-
Structure-function analysis of cytochromes P450 2B.Biochim Biophys Acta. 2007 Mar;1770(3):402-12. doi: 10.1016/j.bbagen.2006.07.006. Epub 2006 Jul 22. Biochim Biophys Acta. 2007. PMID: 16935426 Review.
-
Thermodynamics of ligand binding to P450 2B4 and P450eryF studied by isothermal titration calorimetry.Drug Metab Rev. 2007;39(2-3):539-56. doi: 10.1080/03602530701498182. Drug Metab Rev. 2007. PMID: 17786637 Review.
Cited by
-
Distal effect of amino acid substitutions in CYP2C9 polymorphic variants causes differences in interatomic interactions against (S)-warfarin.PLoS One. 2013 Sep 2;8(9):e74053. doi: 10.1371/journal.pone.0074053. eCollection 2013. PLoS One. 2013. PMID: 24023924 Free PMC article.
-
A review of metabolic and enzymatic engineering strategies for designing and optimizing performance of microbial cell factories.Comput Struct Biotechnol J. 2014 Sep 3;11(18):91-9. doi: 10.1016/j.csbj.2014.08.010. eCollection 2014 Aug. Comput Struct Biotechnol J. 2014. PMID: 25379147 Free PMC article. Review.
-
Functional importance of a peripheral pocket in mammalian cytochrome P450 2B enzymes.Arch Biochem Biophys. 2015 Oct 15;584:61-9. doi: 10.1016/j.abb.2015.08.007. Epub 2015 Aug 28. Arch Biochem Biophys. 2015. PMID: 26319176 Free PMC article.
-
Conformational adaptation of human cytochrome P450 2B6 and rabbit cytochrome P450 2B4 revealed upon binding multiple amlodipine molecules.Biochemistry. 2012 Sep 18;51(37):7225-38. doi: 10.1021/bi300894z. Epub 2012 Sep 4. Biochemistry. 2012. PMID: 22909231 Free PMC article.
-
Steroid-based facial amphiphiles for stabilization and crystallization of membrane proteins.Proc Natl Acad Sci U S A. 2013 Mar 26;110(13):E1203-11. doi: 10.1073/pnas.1221442110. Epub 2013 Mar 11. Proc Natl Acad Sci U S A. 2013. PMID: 23479627 Free PMC article.
References
-
- Williams JA, Hyland R, Jones BC, Smith DA, Hurst S, Goosen TC, Peterkin V, Koup JR, Ball SE. Drug-drug interactions for UDP-glucuronosyltransferase substrates: A pharmacokinetic explanation for typically observed low exposure (AUCi/AUC) ratios. Drug Metab Dispos. 2004;32:1201–1208. - PubMed
-
- Johnson EF, Stout CD. Structural diversity of human xenobiotic-metabolizing cytochrome P450 monooxygenases. Biochem Biophys Res Commun. 2005;338:331–336. - PubMed
-
- Al Omari A, Murry DJ. Pharmacogenetics of the cytochrome P450 enzyme system: Review of current knowledge and clinical significance. J Pharm Pract. 2007;20:206–218.
-
- Guengerich FP. Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem Res Toxicol. 2001;14:611–650. - PubMed
-
- Ingelman-Sundberg M. Human drug metabolising cytochrome P450 enzymes: Properties and polymorphisms. N-S Arch Pharmacol. 2004;369:89–104. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

