Cluster K mycobacteriophages: insights into the evolutionary origins of mycobacteriophage TM4
- PMID: 22053209
- PMCID: PMC3203893
- DOI: 10.1371/journal.pone.0026750
Cluster K mycobacteriophages: insights into the evolutionary origins of mycobacteriophage TM4
Abstract
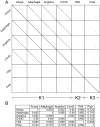
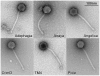
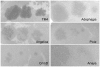
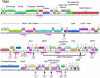
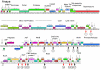
Five newly isolated mycobacteriophages--Angelica, CrimD, Adephagia, Anaya, and Pixie--have similar genomic architectures to mycobacteriophage TM4, a previously characterized phage that is widely used in mycobacterial genetics. The nucleotide sequence similarities warrant grouping these into Cluster K, with subdivision into three subclusters: K1, K2, and K3. Although the overall genome architectures of these phages are similar, TM4 appears to have lost at least two segments of its genome, a central region containing the integration apparatus, and a segment at the right end. This suggests that TM4 is a recent derivative of a temperate parent, resolving a long-standing conundrum about its biology, in that it was reportedly recovered from a lysogenic strain of Mycobacterium avium, but it is not capable of forming lysogens in any mycobacterial host. Like TM4, all of the Cluster K phages infect both fast- and slow-growing mycobacteria, and all of them--with the exception of TM4--form stable lysogens in both Mycobacterium smegmatis and Mycobacterium tuberculosis; immunity assays show that all five of these phages share the same immune specificity. TM4 infects these lysogens suggesting that it was either derived from a heteroimmune temperate parent or that it has acquired a virulent phenotype. We have also characterized a widely-used conditionally replicating derivative of TM4 and identified mutations conferring the temperature-sensitive phenotype. All of the Cluster K phages contain a series of well conserved 13 bp repeats associated with the translation initiation sites of a subset of the genes; approximately one half of these contain an additional sequence feature composed of imperfectly conserved 17 bp inverted repeats separated by a variable spacer. The K1 phages integrate into the host tmRNA and the Cluster K phages represent potential new tools for the genetics of M. tuberculosis and related species.
Conflict of interest statement
Figures










References
-
- Casjens SR. Comparative genomics and evolution of the tailed-bacteriophages. Curr Opin Microbiol. 2005;8:451–458. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

