Improving collision induced dissociation (CID), high energy collision dissociation (HCD), and electron transfer dissociation (ETD) fourier transform MS/MS degradome-peptidome identifications using high accuracy mass information
- PMID: 22054047
- PMCID: PMC3272082
- DOI: 10.1021/pr200597j
Improving collision induced dissociation (CID), high energy collision dissociation (HCD), and electron transfer dissociation (ETD) fourier transform MS/MS degradome-peptidome identifications using high accuracy mass information
Abstract
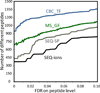
MS dissociation methods, including collision induced dissociation (CID), high energy collision dissociation (HCD), and electron transfer dissociation (ETD), can each contribute distinct peptidome identifications using conventional peptide identification methods (Shen et al. J. Proteome Res. 2011), but such samples still pose significant informatics challenges. In this work, we explored utilization of high accuracy fragment ion mass measurements, in this case provided by Fourier transform MS/MS, to improve peptidome peptide data set size and consistency relative to conventional descriptive and probabilistic scoring methods. For example, we identified 20-40% more peptides than SEQUEST, Mascot, and MS_GF scoring methods using high accuracy fragment ion information and the same false discovery rate (FDR) from CID, HCD, and ETD spectra. Identified species covered >90% of the collective identifications obtained using various conventional peptide identification methods, which significantly addresses the common issue of different data analysis methods generating different peptide data sets. Choice of peptide dissociation and high-precision measurement-based identification methods presently available for degradomic-peptidomic analyses needs to be based on the coverage and confidence (or specificity) afforded by the method, as well as practical issues (e.g., throughput). By using accurate fragment information, >1000 peptidome components can be identified from a single human blood plasma analysis with low peptide-level FDRs (e.g., 0.6%), providing an improved basis for investigating potential disease-related peptidome components.
Figures







References
-
- Walsh CT. Posttranslational modification of proteins. Expanding nature’s inventory. Robert and Company Publishers; Colorado, USA: 2006. Chapter 8.
-
- Edwards D, Høyer-Hansen G, Blasi F, Sloane BF, editors. The cancer degradome: proteases and cancer biology. Springer-verlaq; New York: 2008.
-
- Lópezín-Otín C, Overall CM. Protease degradomics: a new challenge for proteomics. Nat. Rev. Mol. Cell Biol. 2002;4:544–558. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

