More than just lysins: peptidoglycan hydrolases tailor the cell wall
- PMID: 22055466
- PMCID: PMC3347972
- DOI: 10.1016/j.mib.2011.10.003
More than just lysins: peptidoglycan hydrolases tailor the cell wall
Abstract
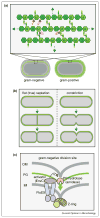
Enzymes that degrade the peptidoglycan (PG) cell wall layer called PG hydrolases or autolysins are often thought of as destructive forces. Phages employ them to lyse their host for the release of virion particles and some bacteria secrete them to eliminate (lyse) their competition. However, bacteria also harness the activity of PG hydrolases for important aspects of growth, division, and development. Of course, using PG hydrolases in this capacity requires that they be tightly regulated. While this has been appreciated for some time, we are only just beginning to understand the mechanisms governing the activities of these 'tailoring' enzymes. This review will focus on recent advances in this area with an emphasis on the regulation of PG hydrolases involved in cell division.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
Similar articles
-
A novel type of peptidoglycan-binding domain highly specific for amidated D-Asp cross-bridge, identified in Lactobacillus casei bacteriophage endolysins.J Biol Chem. 2013 Jul 12;288(28):20416-26. doi: 10.1074/jbc.M112.446344. Epub 2013 Jun 3. J Biol Chem. 2013. PMID: 23733182 Free PMC article.
-
Uncovering the activities, biological roles, and regulation of bacterial cell wall hydrolases and tailoring enzymes.J Biol Chem. 2020 Mar 6;295(10):3347-3361. doi: 10.1074/jbc.REV119.010155. Epub 2020 Jan 23. J Biol Chem. 2020. PMID: 31974163 Free PMC article. Review.
-
Peptidoglycan hydrolases, bacterial shape, and pathogenesis.Curr Opin Microbiol. 2013 Dec;16(6):767-78. doi: 10.1016/j.mib.2013.09.005. Epub 2013 Oct 10. Curr Opin Microbiol. 2013. PMID: 24121030 Review.
-
Control of bacterial cell wall autolysins by peptidoglycan crosslinking mode.Nat Commun. 2024 Sep 11;15(1):7937. doi: 10.1038/s41467-024-52325-2. Nat Commun. 2024. PMID: 39261529 Free PMC article.
-
Bacterial peptidoglycan (murein) hydrolases.FEMS Microbiol Rev. 2008 Mar;32(2):259-86. doi: 10.1111/j.1574-6976.2007.00099.x. Epub 2008 Feb 11. FEMS Microbiol Rev. 2008. PMID: 18266855 Review.
Cited by
-
Harnessing single cell sorting to identify cell division genes and regulators in bacteria.PLoS One. 2013;8(4):e60964. doi: 10.1371/journal.pone.0060964. Epub 2013 Apr 2. PLoS One. 2013. PMID: 23565292 Free PMC article.
-
Two New M23 Peptidoglycan Hydrolases With Distinct Net Charge.Front Microbiol. 2021 Sep 24;12:719689. doi: 10.3389/fmicb.2021.719689. eCollection 2021. Front Microbiol. 2021. PMID: 34630350 Free PMC article.
-
PrkA controls peptidoglycan biosynthesis through the essential phosphorylation of ReoM.Elife. 2020 May 29;9:e56048. doi: 10.7554/eLife.56048. Elife. 2020. PMID: 32469310 Free PMC article.
-
Antibacterial gene transfer across the tree of life.Elife. 2014 Nov 25;3:e04266. doi: 10.7554/eLife.04266. Elife. 2014. PMID: 25422936 Free PMC article.
-
Anti-bacterial glycosyl triazoles - Identification of an N-acetylglucosamine derivative with bacteriostatic activity against Bacillus.Medchemcomm. 2014 Aug;5(8):1213-1217. doi: 10.1039/C4MD00127C. Medchemcomm. 2014. PMID: 25431647 Free PMC article.
References
-
- Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32:149–167. - PubMed
-
- Sauvage E, Kerff F, Terrak M, Ayala J, Charlier P. The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev. 2008;32:234–258. - PubMed
-
- den Blaauwen T, de Pedro MA, Nguyen-Distèche M, Ayala JA. Morphogenesis of rod-shaped sacculi. FEMS Microbiol Rev. 2008;32:321–344. - PubMed
-
- Vollmer W, Joris B, Charlier P, Foster S. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev. 2008;32:259–286. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

