Functional nanoscale organization of signaling molecules downstream of the T cell antigen receptor
- PMID: 22055681
- PMCID: PMC3225724
- DOI: 10.1016/j.immuni.2011.10.004
Functional nanoscale organization of signaling molecules downstream of the T cell antigen receptor
Abstract
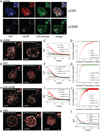
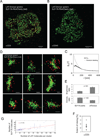
Receptor-regulated cellular signaling often is mediated by formation of transient, heterogeneous protein complexes of undefined structure. We used single and two-color photoactivated localization microscopy to study complexes downstream of the T cell antigen receptor (TCR) in single-molecule detail at the plasma membrane of intact T cells. The kinase ZAP-70 distributed completely with the TCRζ chain and both partially mixed with the adaptor LAT in activated cells, thus showing localized activation of LAT by TCR-coupled ZAP-70. In resting and activated cells, LAT primarily resided in nanoscale clusters as small as dimers whose formation depended on protein-protein and protein-lipid interactions. Surprisingly, the adaptor SLP-76 localized to the periphery of LAT clusters. This nanoscale structure depended on polymerized actin and its disruption affected TCR-dependent cell function. These results extend our understanding of the mechanism of T cell activation and the formation and organization of TCR-mediated signaling complexes, findings also relevant to other receptor systems.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures





References
-
- Ando R, Mizuno H, Miyawaki A. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science. 2004;306:1370–1373. - PubMed
-
- Barda-Saad M, Braiman A, Titerence R, Bunnell SC, Barr VA, Samelson LE. Dynamic molecular interactions linking the T cell antigen receptor to the actin cytoskeleton. Nature Immunology. 2005;6:80–89. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

