Crosstalk between the canonical NF-κB and Notch signaling pathways inhibits Pparγ expression and promotes pancreatic cancer progression in mice
- PMID: 22056382
- PMCID: PMC3225987
- DOI: 10.1172/JCI45797
Crosstalk between the canonical NF-κB and Notch signaling pathways inhibits Pparγ expression and promotes pancreatic cancer progression in mice
Abstract
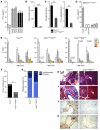
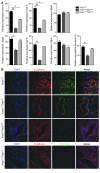
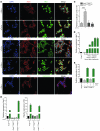
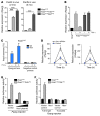
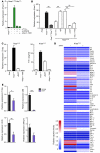
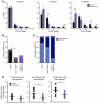
The majority of human pancreatic cancers have activating mutations in the KRAS proto-oncogene. These mutations result in increased activity of the NF-κB pathway and the subsequent constitutive production of proinflammatory cytokines. Here, we show that inhibitor of κB kinase 2 (Ikk2), a component of the canonical NF-κB signaling pathway, synergizes with basal Notch signaling to upregulate transcription of primary Notch target genes, resulting in suppression of antiinflammatory protein expression and promotion of pancreatic carcinogenesis in mice. We found that in the Kras(G12D)Pdx1-cre mouse model of pancreatic cancer, genetic deletion of Ikk2 in initiated pre-malignant epithelial cells substantially delayed pancreatic oncogenesis and resulted in downregulation of the classical Notch target genes Hes1 and Hey1. Tnf-α stimulated canonical NF-κB signaling and, in collaboration with basal Notch signals, induced optimal expression of Notch targets. Mechanistically, Tnf-α stimulation resulted in phosphorylation of histone H3 at the Hes1 promoter, and this signal was lost with Ikk2 deletion. Hes1 suppresses expression of Pparg, which encodes the antiinflammatory nuclear receptor Pparγ. Thus, crosstalk between Tnf-α/Ikk2 and Notch sustains the intrinsic inflammatory profile of transformed cells. These findings reveal what we believe to be a novel interaction between oncogenic inflammation and a major cell fate pathway and show how these pathways can cooperate to promote cancer progression.
Figures


References
-
- Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454(7203):436–444. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

