Repression of the DNA-binding inhibitor Id3 by Blimp-1 limits the formation of memory CD8+ T cells
- PMID: 22057288
- PMCID: PMC3226770
- DOI: 10.1038/ni.2153
Repression of the DNA-binding inhibitor Id3 by Blimp-1 limits the formation of memory CD8+ T cells
Abstract
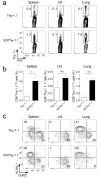
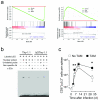
The transcriptional repressor Blimp-1 promotes the differentiation of CD8(+) T cells into short-lived effector cells (SLECs) that express the lectin-like receptor KLRG-1, but how it operates remains poorly defined. Here we show that Blimp-1 bound to and repressed the promoter of the gene encoding the DNA-binding inhibitor Id3 in SLECs. Repression of Id3 by Blimp-1 was dispensable for SLEC development but limited the ability of SLECs to persist as memory cells. Enforced expression of Id3 was sufficient to restore SLEC survival and enhanced recall responses. Id3 function was mediated in part through inhibition of the transcriptional activity of E2A and induction of genes regulating genome stability. Our findings identify the Blimp-1-Id3-E2A axis as a key molecular switch that determines whether effector CD8(+) T cells are programmed to die or enter the memory pool.
Figures






References
-
- Turner CA, Jr., Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77:297–306. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

