Crystal structure of HydF scaffold protein provides insights into [FeFe]-hydrogenase maturation
- PMID: 22057316
- PMCID: PMC3243517
- DOI: 10.1074/jbc.M111.281956
Crystal structure of HydF scaffold protein provides insights into [FeFe]-hydrogenase maturation
Abstract
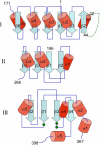
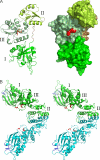
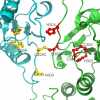
[FeFe]-hydrogenases catalyze the reversible production of H2 in some bacteria and unicellular eukaryotes. These enzymes require ancillary proteins to assemble the unique active site H-cluster, a complex structure composed of a 2Fe center bridged to a [4Fe-4S] cubane. The first crystal structure of a key factor in the maturation process, HydF, has been determined at 3 Å resolution. The protein monomer present in the asymmetric unit of the crystal comprises three domains: a GTP-binding domain, a dimerization domain, and a metal cluster-binding domain, all characterized by similar folding motifs. Two monomers dimerize, giving rise to a stable dimer, held together mainly by the formation of a continuous β-sheet comprising eight β-strands from two monomers. Moreover, in the structure presented, two dimers aggregate to form a supramolecular organization that represents an inactivated form of the HydF maturase. The crystal structure of the latter furnishes several clues about the events necessary for cluster generation/transfer and provides an excellent model to begin elucidating the structure/function of HydF in [FeFe]-hydrogenase maturation.
Figures
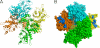
References
-
- Vignais P. M., Billoud B. (2007) Chem. Rev. 107, 4206–4272 - PubMed
-
- Ghirardi M. L., Posewitz M. C., Maness P. C., Dubini A., Yu J., Seibert M. (2007) Annu. Rev. Plant. Biol. 58, 71–91 - PubMed
-
- Adams M. W. (1990) Biochim. Biophys. Acta 1020, 115–145 - PubMed
-
- Peters J. W., Lanzilotta W. N., Lemon B. J., Seefeldt L. C. (1998) Science 282, 1853–1858 - PubMed
-
- Nicolet Y., Piras C., Legrand P., Hatchikian C. E., Fontecilla-Camps J. C. (1999) Structure 7, 13–23 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical

