pre-miRNA profiles obtained through application of locked nucleic acids and deep sequencing reveals complex 5'/3' arm variation including concomitant cleavage and polyuridylation patterns
- PMID: 22058130
- PMCID: PMC3287202
- DOI: 10.1093/nar/gkr903
pre-miRNA profiles obtained through application of locked nucleic acids and deep sequencing reveals complex 5'/3' arm variation including concomitant cleavage and polyuridylation patterns
Abstract
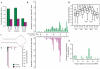
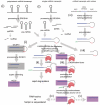
Recent research hints at an underappreciated complexity in pre-miRNA processing and regulation. Global profiling of pre-miRNA and its potential to increase understanding of the pre-miRNA landscape is impeded by overlap with highly expressed classes of other non coding (nc) RNA. Here, we present a data set excluding these RNA before sequencing through locked nucleic acids (LNA), greatly increasing pre-miRNA sequence counts with no discernable effect on pre-miRNA or mature miRNA sequencing. Analysis of profiles generated in total, nuclear and cytoplasmic cell fractions reveals that pre-miRNAs are subject to a wide range of regulatory processes involving loci-specific 3'- and 5'-end variation entailing complex cleavage patterns with co-occurring polyuridylation. Additionally, examination of nuclear-enriched flanking sequences of pre-miRNA, particularly those derived from polycistronic miRNA transcripts, provides insight into miRNA and miRNA-offset (moRNA) production, specifically identifying novel classes of RNA potentially functioning as moRNA precursors. Our findings point to particularly intricate regulation of the let-7 family in many ways reminiscent of DICER1-independent, pre-mir-451-like processing, introduce novel and unify known forms of pre-miRNA regulation and processing, and shed new light on overlooked products of miRNA processing pathways.
Figures




References
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. - PubMed
-
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. - PubMed
-
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. - PubMed
-
- Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, Yoneda Y, Tsukihara T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science. 2009;326:1275–1279. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

