Sulfoquinovose synthase - an important enzyme in the N-glycosylation pathway of Sulfolobus acidocaldarius
- PMID: 22059775
- PMCID: PMC4391026
- DOI: 10.1111/j.1365-2958.2011.07875.x
Sulfoquinovose synthase - an important enzyme in the N-glycosylation pathway of Sulfolobus acidocaldarius
Abstract
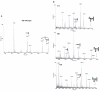
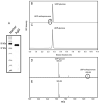
Recently, the Surface (S)-layer glycoprotein of the thermoacidophilic crenarchaeote Sulfolobus acidocaldarius was found to be N-glycosylated with a heterogeneous family of glycans, with the largest having a composition Glc(1)Man(2)GlcNAc(2) plus 6-sulfoquinovose. However, genetic analyses of genes involved in the N-glycosylation process in Crenarchaeota were missing so far. In this study we identify a gene cluster involved in the biosynthesis of sulfoquinovose and important for the assembly of the S-layer N-glycans. A successful markerless in-frame deletion of agl3 resulted in a decreased molecular mass of the S-layer glycoprotein SlaA and the flagellin FlaB, indicating a change in the N-glycan composition. Analyses with nanoLC ES-MS/MS confirmed the presence of only a reduced trisaccharide structure composed of Man(1) GlcNAc(2) , missing the sulfoquinovose, a mannose and glucose. Biochemical studies of the recombinant Agl3 confirmed the proposed function as a UDP-sulfoquinovose synthase. Furthermore, S. acidocaldarius cells lacking agl3 had a significantly lower growth rate at elevated salt concentrations compared with the background strain, underlining the importance of the N-glycosylation to maintain an intact and stable cell envelope, to enable the survival of S. acidocaldarius in its extreme environment.
© 2011 Blackwell Publishing Ltd.
Figures




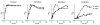

References
-
- Abu-Qarn M, Yurist-Doutsch S, Giordano A, Trauner A, Morris HR, Hitchen P, et al. Haloferax volcanii AglB and AglD are involved in N-glycosylation of the S-layer glycoprotein and proper assembly of the surface layer. J Mol Biol. 2007;374:1224–1236. - PubMed
-
- Albers SV, Meyer BH. The archaeal cell envelope. Nat Rev Microbiol. 2011;9:414–426. - PubMed
-
- Benning C. Biosynthesis and function of the sulfolipid sulfoquinovosyl diacylglycerol. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:53–75. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

