Direct evidence of nuclear Argonaute distribution during transcriptional silencing links the actin cytoskeleton to nuclear RNAi machinery in human cells
- PMID: 22064859
- PMCID: PMC3287199
- DOI: 10.1093/nar/gkr891
Direct evidence of nuclear Argonaute distribution during transcriptional silencing links the actin cytoskeleton to nuclear RNAi machinery in human cells
Abstract
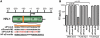
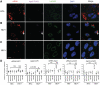
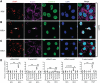
Mammalian RNAi machinery facilitating transcriptional gene silencing (TGS) is the RNA-induced transcriptional gene silencing-like (RITS-like) complex, comprising of Argonaute (Ago) and small interfering RNA (siRNA) components. We have previously demonstrated promoter-targeted siRNA induce TGS in human immunodeficiency virus type-1 (HIV-1) and simian immunodeficiency virus (SIV), which profoundly suppresses retrovirus replication via heterochromatin formation and histone methylation. Here, we examine subcellular co-localization of Ago proteins with promoter-targeted siRNAs during TGS of SIV and HIV-1 infection. Analysis of retrovirus-infected cells revealed Ago1 co-localized with siRNA in the nucleus, while Ago2 co-localized with siRNA in the inner nuclear envelope. Mismatched and scrambled siRNAs were observed in the cytoplasm, indicating sequence specificity. This is the first report directly visualizing nuclear compartment distribution of Ago-associated siRNA and further reveals a novel nuclear trafficking mechanism for RITS-like components involving the actin cytoskeleton. These results establish a model for elucidating mammalian TGS and suggest a fundamental mechanism underlying nuclear delivery of RITS-like components.
Figures
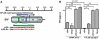





References
-
- Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat. Struct. Mol. Biol. 2006;13:793–797. - PubMed
-
- Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. - PubMed
-
- Robb GB, Brown KM, Khurana J, Rana TM. Specific and potent RNAi in the nucleus of human cells. Nat. Struct. Mol. Biol. 2005;12:133–137. - PubMed
-
- Lim HG, Suzuki K, Cooper DA, Kelleher AD. Promoter-targeted siRNAs induce gene silencing of simian immunodeficiency virus (SIV) infection in vitro. Mol. Ther. 2008;16:565–570. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

