Formation of ternary complex of human biliverdin reductase-protein kinase Cδ-ERK2 protein is essential for ERK2-mediated activation of Elk1 protein, nuclear factor-κB, and inducible nitric-oxidase synthase (iNOS)
- PMID: 22065579
- PMCID: PMC3256887
- DOI: 10.1074/jbc.M111.279612
Formation of ternary complex of human biliverdin reductase-protein kinase Cδ-ERK2 protein is essential for ERK2-mediated activation of Elk1 protein, nuclear factor-κB, and inducible nitric-oxidase synthase (iNOS)
Abstract
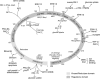
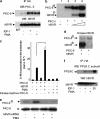
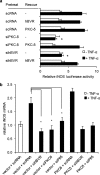
Growth factors, insulin, oxidative stress, and cytokines activate ERK1/2 by PKCδ and MEK1/2. Human biliverdin reductase (hBVR), a Ser/Thr/Tyr kinase and intracellular scaffold/bridge/anchor, is a nuclear transporter of MEK1/2-stimulated ERK1/2 (Lerner-Marmarosh, N., Miralem, T., Gibbs, P. E., and Maines, M. D. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6870-6875). hBVR, PKCδ, and MEK1/2 overlap in their tissue expression profile and type of activators. Presently, we report on formation of an hBVR-PKCδ-ERK2 ternary complex that is essential for ERK2 signal transduction and activation of genes linked to cell proliferation and cancer. MEK1/2 and the protein phosphatase PP2A were also present in the complex. When cells were stimulated with insulin-like growth factor-1 (IGF-1), an increased interaction between hBVR and PKCδ was detected by FRET-fluorescence lifetime imaging microscopy. hBVR and ERK2 were phosphorylated by PKCδ; however, the PKC was not a substrate for either ERK2 or hBVR. IGF-1 and phorbol ester increased hBVR/PKCδ binding; hBVR was required for the activation of PKCδ and its interaction with ERK2. The C-terminal phenylalanine residues of PKCδ (Phe(660), Phe(663), and Phe(665)) were necessary for binding to ERK2 but not for hBVR binding. Formation of the hBVR-PKCδ-ERK2 complex required the hBVR docking site for ERK, FXFP (DEF, C-box) and D(δ)-box (ILXXLXL) motifs. The hBVR-based peptide KKRILHCLGLA inhibited PKC activation and PKCδ/ERK2 interaction. Phorbol ester- and TNF-α-dependent activation of the ERK-regulated transcription factors Elk1 and NF-κB and expression of the iNOS gene were suppressed by hBVR siRNA; those activities were rescued by hBVR. The findings reveal the direct input of hBVR in PKCδ/ERK signaling and identify hBVR-based peptide regulators of ERK-mediated gene activation.
Figures







References
-
- Jackson D. N., Foster D. A. (2004) FASEB J. 18, 627–636 - PubMed
-
- Boutros T., Chevet E., Metrakos P. (2008) Pharmacol. Rev. 60, 261–310 - PubMed
-
- Kim E. K., Choi E. J. (2010) Biochim. Biophys. Acta 1802, 396–405 - PubMed
-
- Ranganathan A., Yazicioglu M. N., Cobb M. H. (2006) J. Biol. Chem. 281, 15645–15652 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

