Polypeptide modulators of caspase recruitment domain (CARD)-CARD-mediated protein-protein interactions
- PMID: 22065589
- PMCID: PMC3247988
- DOI: 10.1074/jbc.M111.255364
Polypeptide modulators of caspase recruitment domain (CARD)-CARD-mediated protein-protein interactions
Abstract
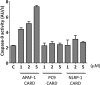
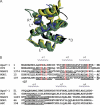
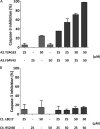
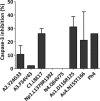
The caspase recruitment domain (CARD) is present in a large number of proteins. Initially, the CARD was recognized as part of the caspase activation machinery. CARD-CARD interactions play a role in apoptosis and are responsible for the Apaf-1-mediated activation of procaspase-9 in the apoptosome. CARD-containing proteins mediate the inflammasome-dependent activation of proinflammatory caspase-1. More recently, new roles for CARD-containing proteins have been reported in signaling pathways associated with immune responses. The functional role of CARD-containing proteins and CARDs in coordinating apoptosis and inflammatory and immune responses is not completely understood. We have explored the putative cross-talk between apoptosis and inflammation by analyzing the modulatory activity on both the Apaf-1/procaspase-9 interaction and the inflammasome-mediated procaspase-1 activation of CARD-derived polypeptides. To this end, we analyzed the activity of individual recombinant CARDs, rationally designed CARD-derived peptides, and peptides derived from phage display.
Figures



Similar articles
-
The Apaf-1*procaspase-9 apoptosome complex functions as a proteolytic-based molecular timer.EMBO J. 2009 Jul 8;28(13):1916-25. doi: 10.1038/emboj.2009.152. Epub 2009 Jun 4. EMBO J. 2009. PMID: 19494828 Free PMC article.
-
Mechanistic insights into caspase-9 activation by the structure of the apoptosome holoenzyme.Proc Natl Acad Sci U S A. 2017 Feb 14;114(7):1542-1547. doi: 10.1073/pnas.1620626114. Epub 2017 Jan 31. Proc Natl Acad Sci U S A. 2017. PMID: 28143931 Free PMC article.
-
Apoptosis-associated speck-like protein containing a caspase recruitment domain is a regulator of procaspase-1 activation.J Immunol. 2003 Dec 1;171(11):6154-63. doi: 10.4049/jimmunol.171.11.6154. J Immunol. 2003. PMID: 14634131
-
New insights into apoptosome structure and function.Cell Death Differ. 2018 Jul;25(7):1194-1208. doi: 10.1038/s41418-017-0025-z. Epub 2018 May 15. Cell Death Differ. 2018. PMID: 29765111 Free PMC article. Review.
-
Apoptosome structure, assembly, and procaspase activation.Structure. 2013 Apr 2;21(4):501-15. doi: 10.1016/j.str.2013.02.024. Structure. 2013. PMID: 23561633 Free PMC article. Review.
Cited by
-
Computational Characterization of the Interaction of CARD Domains in the Apoptosome.Biochemistry. 2025 Jan 21;64(2):401-418. doi: 10.1021/acs.biochem.4c00583. Epub 2025 Jan 6. Biochemistry. 2025. PMID: 39761026 Free PMC article.
-
Spontaneous and transgenic rodent models of inflammatory bowel disease.Lab Anim Res. 2015 Jun;31(2):47-68. doi: 10.5625/lar.2015.31.2.47. Epub 2015 Jun 26. Lab Anim Res. 2015. PMID: 26155200 Free PMC article. Review.
-
Mild microwave ablation combined with HSP90 and TGF‑β1 inhibitors enhances the therapeutic effect on osteosarcoma.Mol Med Rep. 2020 Aug;22(2):906-914. doi: 10.3892/mmr.2020.11173. Epub 2020 May 22. Mol Med Rep. 2020. PMID: 32468060 Free PMC article.
-
Kaposi's sarcoma: a computational approach through protein-protein interaction and gene regulatory networks analysis.Virus Genes. 2013 Apr;46(2):242-54. doi: 10.1007/s11262-012-0865-z. Epub 2012 Dec 25. Virus Genes. 2013. PMID: 23266878
-
Mechanisms of Caspases 3/7/8/9 in the Degeneration of External Gills of Chinese Giant Salamanders (Andrias davidianus).Genes (Basel). 2022 Jul 29;13(8):1360. doi: 10.3390/genes13081360. Genes (Basel). 2022. PMID: 36011271 Free PMC article.
References
-
- Seth R. B., Sun L., Ea C. K., Chen Z. J. (2005) Cell 122, 669-682 - PubMed
-
- Hiscott J., Lin R., Nakhaei P., Paz S. (2006) Trends Mol. Med. 12, 53–56 - PubMed
-
- Inohara N., Koseki T., del Peso L., Hu Y., Yee C., Chen S., Carrio R., Merino J., Liu D., Ni J., Núñez G. (1999) J. Biol. Chem. 274, 14560–14567 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

