Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae
- PMID: 22065637
- PMCID: PMC3257523
- DOI: 10.1083/jcb.201106037
Hsp42 is required for sequestration of protein aggregates into deposition sites in Saccharomyces cerevisiae
Abstract
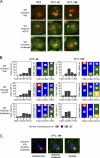
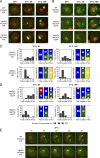
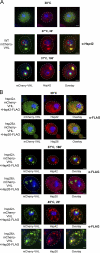
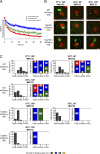
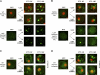
The aggregation of proteins inside cells is an organized process with cytoprotective function. In Saccharomyces cerevisiae, aggregating proteins are spatially sequestered to either juxtanuclear or peripheral sites, which target distinct quality control pathways for refolding and degradation. The cellular machinery driving the sequestration of misfolded proteins to these sites is unknown. In this paper, we show that one of the two small heat shock proteins of yeast, Hsp42, is essential for the formation of peripheral aggregates during physiological heat stress. Hsp42 preferentially localizes to peripheral aggregates but is largely absent from juxtanuclear aggregates, which still form in hsp42Δ cells. Transferring the amino-terminal domain of Hsp42 to Hsp26, which does not participate in aggregate sorting, enables Hsp26 to replace Hsp42 function. Our data suggest that Hsp42 acts via its amino-terminal domain to coaggregate with misfolded proteins and perhaps link such complexes to further sorting factors.
© 2011 Specht et al.
Figures


References
-
- Abelson J.N., Guthrie C., Fink G.R. 2003. Guide to Yeast Genetics and Molecular Biology. Academic Press, New York: 933 pp
-
- Arrigo A.P. 2000. sHsp as novel regulators of programmed cell death and tumorigenicity. Pathol. Biol. (Paris). 48:280–288 - PubMed
-
- Ayscough K.R., Stryker J., Pokala N., Sanders M., Crews P., Drubin D.G. 1997. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 137:399–416 10.1083/jcb.137.2.399 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

