Metabolic cycling without cell division cycling in respiring yeast
- PMID: 22065748
- PMCID: PMC3223432
- DOI: 10.1073/pnas.1116998108
Metabolic cycling without cell division cycling in respiring yeast
Abstract
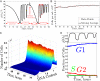
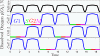
Despite rapid progress in characterizing the yeast metabolic cycle, its connection to the cell division cycle (CDC) has remained unclear. We discovered that a prototrophic batch culture of budding yeast, growing in a phosphate-limited ethanol medium, synchronizes spontaneously and goes through multiple metabolic cycles, whereas the fraction of cells in the G1/G0 phase of the CDC increases monotonically from 90 to 99%. This demonstrates that metabolic cycling does not require cell division cycling and that metabolic synchrony does not require carbon-source limitation. More than 3,000 genes, including most genes annotated to the CDC, were expressed periodically in our batch culture, albeit a mere 10% of the cells divided asynchronously; only a smaller subset of CDC genes correlated with cell division. These results suggest that the yeast metabolic cycle reflects a growth cycle during G1/G0 and explains our previous puzzling observation that genes annotated to the CDC increase in expression at slow growth.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Küenzi MT, Fiechter A. Changes in carbohydrate composition and trehalase-activity during the budding cycle of Saccharomyces cerevisiae. Arch Mikrobiol. 1969;64:396–407. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

