Insights regarding guanine nucleotide exchange from the structure of a DENN-domain protein complexed with its Rab GTPase substrate
- PMID: 22065758
- PMCID: PMC3219131
- DOI: 10.1073/pnas.1110415108
Insights regarding guanine nucleotide exchange from the structure of a DENN-domain protein complexed with its Rab GTPase substrate
Abstract
Rab GTPases are key regulators of membrane traffic pathways within eukaryotic cells. They are specifically activated by guanine nucleotide exchange factors (GEFs), which convert them from their "inactive" GDP-bound form to the "active" GTP-bound form. In higher eukaryotes, proteins containing DENN-domains comprise a major GEF family. Here we describe at 2.1-Å resolution the first structure of a DENN-domain protein, DENND1B-S, complexed with its substrate Rab35, providing novel insights as to how DENN-domain GEFs interact with and activate Rabs. DENND1B-S is bi-lobed, and interactions with Rab35 are through conserved surfaces in both lobes. Rab35 binds via switch regions I and II, around the nucleotide-binding pocket. Positional shifts in Rab residues required for nucleotide binding may lower its affinity for bound GDP, and a conformational change in switch I, which makes the nucleotide-binding pocket more solvent accessible, likely also facilitates exchange.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
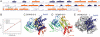
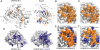

References
-
- Cai H, Reinisch K, Ferro-Novick S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell. 2007;12:671–682. - PubMed
-
- Behnia R, Munro S. Organelle identity and the signposts for membrane traffic. Nature. 2005;438:597–604. - PubMed
-
- Pfeffer SR, Aivazian D. Targeting Rab GTPases to distinct membrane compartments. Nat Rev Mol Cell Biol. 2004;5:886–896. - PubMed
-
- Zerial M, McBride H. Rab proteins as membrane organizers. 2001;2:107–117. - PubMed
-
- Goody RS, Hofmann-Goody W. Exchange factors, effectors, GAPs and motor proteins: Common thermodynamic and kinetic principles for different functions. Eur Biophys J. 2002;31:268–274. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

