Base-flipping mechanism in postmismatch recognition by MutS
- PMID: 22067162
- PMCID: PMC3207177
- DOI: 10.1016/j.bpj.2011.09.045
Base-flipping mechanism in postmismatch recognition by MutS
Abstract
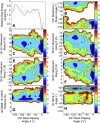
DNA mismatch recognition and repair is vital for preserving the fidelity of the genome. Conserved across prokaryotes and eukaryotes, MutS is the primary protein that is responsible for recognizing a variety of DNA mismatches. From molecular dynamics simulations of the Escherichia coli MutS-DNA complex, we describe significant conformational dynamics in the DNA surrounding a G·T mismatch that involves weakening of the basepair hydrogen bonding in the basepair adjacent to the mismatch and, in one simulation, complete base opening via the major groove. The energetics of base flipping was further examined with Hamiltonian replica exchange free energy calculations revealing a stable flipped-out state with an initial barrier of ~2 kcal/mol. Furthermore, we observe changes in the local DNA structure as well as in the MutS structure that appear to be correlated with base flipping. Our results suggest a role of base flipping as part of the repair initiation mechanism most likely leading to sliding-clamp formation.
Copyright © 2011 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
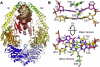

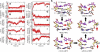
References
-
- Lahue R.S., Au K.G., Modrich P. DNA mismatch correction in a defined system. Science. 1989;245:160–164. - PubMed
-
- Grilley M., Griffith J., Modrich P. Bidirectional excision in methyl-directed mismatch repair. J. Biol. Chem. 1993;268:11830–11837. - PubMed
-
- Cooper D.L., Lahue R.S., Modrich P. Methyl-directed mismatch repair is bidirectional. J. Biol. Chem. 1993;268:11823–11829. - PubMed
-
- Au K.G., Welsh K., Modrich P. Initiation of methyl-directed mismatch repair. J. Biol. Chem. 1992;267:12142–12148. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

