Epigenetic-aging-signature to determine age in different tissues
- PMID: 22067257
- PMCID: PMC3229965
- DOI: 10.18632/aging.100395
Epigenetic-aging-signature to determine age in different tissues
Abstract
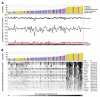
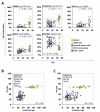
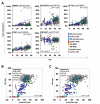
All tissues of the organism are affected by aging. This process is associated with epigenetic modifications such as methylation changes at specific cytosine residues in the DNA (CpG sites). Here, we have identified an Epigenetic-Aging-Signature which is applicable for many tissues to predict donor age. DNA-methylation profiles of various cell types were retrieved from public data depositories - all using the HumanMethylation27 BeadChip platform which represents 27,578 CpG sites. Five datasets from dermis, epidermis, cervical smear, T-cells and monocytes were used for Pavlidis Template Matching to identify 19 CpG sites that are continuously hypermethylated upon aging (R>0.6; p-value<10-13). Four of these CpG sites (associated with the genes NPTX2, TRIM58, GRIA2 and KCNQ1DN) and an additional hypomethylated CpG site (BIRC4BP) were implemented in a model to predict donor age. This Epigenetic-Aging-Signature was tested on a validation group of eight independent datasets corresponding to several cell types from different tissues. Overall, the five CpG sites revealed age-associated DNA-methylation changes in all tissues. The average absolute difference between predicted and real chronological age was about 11 years. This method can be used to predict donor age in various cell preparations - for example in forensic analysis.
Conflict of interest statement
The authors of this manuscript have no conflict of interest to declare.
Figures

References
-
- Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23:413–418. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

