Molecular characterisation of formalin-fixed paraffin-embedded (FFPE) breast tumour specimens using a custom 512-gene breast cancer bead array-based platform
- PMID: 22067903
- PMCID: PMC3242517
- DOI: 10.1038/bjc.2011.355
Molecular characterisation of formalin-fixed paraffin-embedded (FFPE) breast tumour specimens using a custom 512-gene breast cancer bead array-based platform
Abstract
Background: Formalin-fixed, paraffin-embedded (FFPE) tumour tissue represents an immense but mainly untapped resource with respect to molecular profiling. The DASL (cDNA-mediated Annealing, Selection, extension, and Ligation) assay is a recently described, RT-PCR-based, highly multiplexed high-throughput gene expression platform developed by Illumina specifically for fragmented RNA typically obtained from FFPE specimens, which enables expression profiling. In order to extend the utility of the DASL assay for breast cancer, we have custom designed and validated a 512-gene human breast cancer panel.
Methods: The RNA from FFPE breast tumour specimens were analysed using the DASL assay. Breast cancer subtype was defined from pathology immunohistochemical (IHC) staining. Differentially expressed genes between the IHC-defined subtypes were assessed by prediction analysis of microarrays (PAM) and then used in the analysis of two published data sets with clinical outcome data.
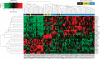
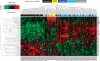
Results: Gene expression signatures on our custom breast cancer panel were very reproducible between replicates (average Pearson's R²=0.962) and the 152 genes common to both the standard cancer DASL panel (Illumina) and our breast cancer DASL panel were similarly expressed for samples run on both panels (average R²=0.877). Moreover, expression of ESR1, PGR and ERBB2 corresponded well with their respective pathology-defined IHC status. A 30-gene set indicative of IHC-defined breast cancer subtypes was found to segregate samples based on their subtype in our data sets and published data sets. Furthermore, several of these genes were significantly associated with overall survival (OS) and relapse-free survival (RFS) in these previously published data sets, indicating that they are biomarkers of the different breast cancer subtypes and the prognostic outcomes associated with these subtypes.
Conclusion: We have demonstrated the ability to expression profile degraded RNA transcripts derived from FFPE tissues on the DASL platform. Importantly, we have identified a 30-biomarker gene set that can classify breast cancer into subtypes and have shown that a subset of these markers is prognostic of OS and RFS.
2011 Cancer Research UK
Figures

References
-
- Arriola E, Marchio C, Tan DS, Drury SC, Lambros MB, Natrajan R, Rodriguez-Pinilla SM, Mackay A, Tamber N, Fenwick K, Jones C, Dowsett M, Ashworth A, Reis-Filho JS (2008) Genomic analysis of the HER2/TOP2A amplicon in breast cancer and breast cancer cell lines. Lab Invest 88: 491–503 - PubMed
-
- Ayers M, Symmans WF, Stec J, Damokosh AI, Clark E, Hess K, Lecocke M, Metivier J, Booser D, Ibrahim N, Valero V, Royce M, Arun B, Whitman G, Ross J, Sneige N, Hortobagyi GN, Pusztai L (2004) Gene expression profiles predict complete pathologic response to neoadjuvant paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide chemotherapy in breast cancer. J Clin Oncol 22: 2284–2293 - PubMed
-
- Bibikova M, Chudin E, Arsanjani A, Zhou L, Garcia EW, Modder J, Kostelec M, Barker D, Downs T, Fan JB, Wang-Rodriguez J (2007) Expression signatures that correlated with Gleason score and relapse in prostate cancer. Genomics 89: 666–672 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

