EZH2 Mediates epigenetic silencing of neuroblastoma suppressor genes CASZ1, CLU, RUNX3, and NGFR
- PMID: 22068036
- PMCID: PMC3487161
- DOI: 10.1158/0008-5472.CAN-11-0961
EZH2 Mediates epigenetic silencing of neuroblastoma suppressor genes CASZ1, CLU, RUNX3, and NGFR
Abstract
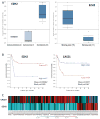
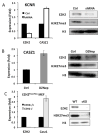
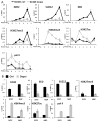
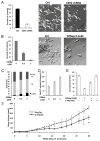
Neuroblastoma (NB) is the most common extracranial pediatric solid tumor with an undifferentiated status and generally poor prognosis, but the basis for these characteristics remains unknown. In this study, we show that upregulation of the Polycomb protein histone methyltransferase EZH2, which limits differentiation in many tissues, is critical to maintain the undifferentiated state and poor prognostic status of NB by epigenetic repression of multiple tumor suppressor genes. We identified this role for EZH2 by examining the regulation of CASZ1, a recently identified NB tumor suppressor gene whose ectopic restoration inhibits NB cell growth and induces differentiation. Reducing EZH2 expression by RNA interference-mediated knockdown or pharmacologic inhibiton with 3-deazaneplanocin A increased CASZ1 expression, inhibited NB cell growth, and induced neurite extension. Similarly, EZH2(-/-) mouse embryonic fibroblasts (MEF) displayed 3-fold higher levels of CASZ1 mRNA compared with EZH2(+/+) MEFs. In cells with increased expression of CASZ1, treatment with histone deacetylase (HDAC) inhibitor decreased expression of EZH2 and the Polycomb Repressor complex component SUZ12. Under steady-state conditions, H3K27me3 and PRC2 components bound to the CASZ1 gene were enriched, but this enrichment was decreased after HDAC inhibitor treatment. We determined that the tumor suppressors CLU, NGFR, and RUNX3 were also directly repressed by EZH2 like CASZ1 in NB cells. Together, our findings establish that aberrant upregulation of EZH2 in NB cells silences several tumor suppressors, which contribute to the genesis and maintenance of the undifferentiated phenotype of NB tumors.
©2011 AACR.
Figures



References
-
- Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–20. - PubMed
-
- Oberthuer A, Theissen J, Westermann F, Hero B, Fischer M. Molecular characterization and classification of neuroblastoma. Future Oncol. 2009;5:625–39. - PubMed
-
- Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–16. - PubMed
-
- Krasnoselsky AL, Whiteford CC, Wei JS, Bilke S, Westermann F, Chen QR, et al. Altered expression of cell cycle genes distinguishes aggressive neuroblastoma. Oncogene. 2005;24:1533–41. - PubMed
-
- Caron H, van Sluis P, de Kraker J, Bokkerink J, Egeler M, Laureys G, et al. Allelic loss of chromosome 1p as a predictor of unfavorable outcome in patients with neuroblastoma. N Engl J Med. 1996;334:225–30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

