The quantitative proteome of a human cell line
- PMID: 22068332
- PMCID: PMC3261713
- DOI: 10.1038/msb.2011.82
The quantitative proteome of a human cell line
Abstract
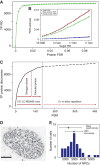
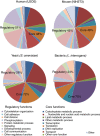
The generation of mathematical models of biological processes, the simulation of these processes under different conditions, and the comparison and integration of multiple data sets are explicit goals of systems biology that require the knowledge of the absolute quantity of the system's components. To date, systematic estimates of cellular protein concentrations have been exceptionally scarce. Here, we provide a quantitative description of the proteome of a commonly used human cell line in two functional states, interphase and mitosis. We show that these human cultured cells express at least -10 000 proteins and that the quantified proteins span a concentration range of seven orders of magnitude up to 20 000 000 copies per cell. We discuss how protein abundance is linked to function and evolution.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 - PMC - PubMed
-
- Bodenmiller B, Wanka S, Kraft C, Urban J, Campbell D, Pedrioli PG, Gerrits B, Picotti P, Lam H, Vitek O, Brusniak MY, Roschitzki B, Zhang C, Shokat KM, Schlapbach R, Colman-Lerner A, Nolan GP, Nesvizhskii AI, Peter M, Loewith R et al. (2010) Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci Signal 3: rs4. - PMC - PubMed
-
- Claassen M, Aebersold R, Buhmann JM (2011) Proteome coverage prediction for integrated proteomics datasets. J Comput Biol 18: 283–293 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

