A monomorphic haplotype of chromosome Ia is associated with widespread success in clonal and nonclonal populations of Toxoplasma gondii
- PMID: 22068979
- PMCID: PMC3215432
- DOI: 10.1128/mBio.00228-11
A monomorphic haplotype of chromosome Ia is associated with widespread success in clonal and nonclonal populations of Toxoplasma gondii
Abstract
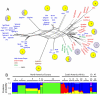
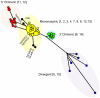
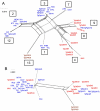
Toxoplasma gondii is a common parasite of animals that also causes a zoonotic infection in humans. Previous studies have revealed a strongly clonal population structure that is shared between North America and Europe, while South American strains show greater genetic diversity and evidence of sexual recombination. The common inheritance of a monomorphic version of chromosome Ia (referred to as ChrIa*) among three clonal lineages from North America and Europe suggests that inheritance of this chromosome might underlie their recent clonal expansion. To further examine the diversity and distribution of ChrIa, we have analyzed additional strains with greater geographic diversity. Our findings reveal that the same haplotype of ChrIa* is found in the clonal lineages from North America and Europe and in older lineages in South America, where sexual recombination is more common. Although lineages from all three continents harbor the same conserved ChrIa* haplotype, strains from North America and Europe are genetically separate from those in South America, and these respective geographic regions show limited evidence of recent mixing. Genome-wide, array-based profiling of polymorphisms provided evidence for an ancestral flow from particular older southern lineages that gave rise to the clonal lineages now dominant in the north. Collectively, these data indicate that ChrIa* is widespread among nonclonal strains in South America and has more recently been associated with clonal expansion of specific lineages in North America and Europe. These findings have significant implications for the spread of genetic loci influencing transmission and virulence in pathogen populations.
Importance: Understanding parasite population structure is important for evaluating the potential spread of pathogenicity determinants between different geographic regions. Examining the genetic makeup of different isolates of Toxoplasma gondii from around the world revealed that chromosome Ia is highly homogeneous among lineages that predominate on different continents and within genomes that were otherwise quite divergent. This pattern of recent shared ancestry is highly unusual and suggests that some gene(s) found on this chromosome imparts an unusual fitness advantage that has resulted in its recent spread. Although the basis for the conservation of this particularly homogeneous chromosome is unknown, it may have implications for the transmission of infection and spread of human disease.
Figures


References
-
- Dubey JP. 2010. Toxoplasmosis of animals and humans. CRC Press, Boca Raton, FL
-
- Joynson DH, Wreghitt TJ. 2001. Toxoplasmosis: a comprehensive clinical guide. Cambridge University Press, Cambridge, United Kingdom
-
- Benenson MW, Takafuji ET, Lemon SM, Greenup RL, Sulzer AJ. 1982. Oocyst-transmitted toxoplasmosis associated with ingestion of contaminated water. N. Engl. J. Med. 307:666–669 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
