An inducible cell-cell fusion system with integrated ability to measure the efficiency and specificity of HIV-1 entry inhibitors
- PMID: 22069466
- PMCID: PMC3206054
- DOI: 10.1371/journal.pone.0026731
An inducible cell-cell fusion system with integrated ability to measure the efficiency and specificity of HIV-1 entry inhibitors
Abstract
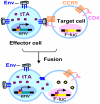
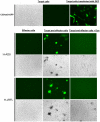
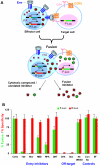
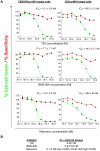
HIV-1 envelope glycoproteins (Envs) mediate virus entry by fusing the viral and target cell membranes, a multi-step process that represents an attractive target for inhibition. Entry inhibitors with broad-range activity against diverse isolates of HIV-1 may be extremely useful as lead compounds for the development of therapies or prophylactic microbicides. To facilitate the identification of such inhibitors, we have constructed a cell-cell fusion system capable of simultaneously monitoring inhibition efficiency and specificity. In this system, effector cells stably express a tetracycline-controlled transactivator (tTA) that enables tightly inducible expression of both HIV-1 Env and the Renilla luciferase (R-Luc) reporter protein. Target cells express the HIV-1 receptors, CD4 and CCR5, and carry the firefly luciferase (F-Luc) reporter gene under the control of a tTA-responsive promoter. Thus, Env-mediated fusion of these two cell types allows the tTA to diffuse to the target cell and activate the expression of the F-Luc protein. The efficiency with which an inhibitor blocks cell-cell fusion is measured by a decrease in the F-Luc activity, while the specificity of the inhibitor is evaluated by its effect on the R-Luc activity. The system exhibited a high dynamic range and high Z'-factor values. The assay was validated with a reference panel of inhibitors that target different steps in HIV-1 entry, yielding inhibitory concentrations comparable to published virus inhibition data. Our system is suitable for large-scale screening of chemical libraries and can also be used for detailed characterization of inhibitory and cytotoxic properties of known entry inhibitors.
Conflict of interest statement
Figures


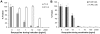

Similar articles
-
Characterization of HIV-1 entry inhibitors with broad activity against R5 and X4 viral strains.J Transl Med. 2015 Apr 2;13:107. doi: 10.1186/s12967-015-0461-9. J Transl Med. 2015. PMID: 25888743 Free PMC article.
-
Activation and Inactivation of Primary Human Immunodeficiency Virus Envelope Glycoprotein Trimers by CD4-Mimetic Compounds.J Virol. 2017 Jan 18;91(3):e01880-16. doi: 10.1128/JVI.01880-16. Print 2017 Feb 1. J Virol. 2017. PMID: 27881646 Free PMC article.
-
HIV-1 predisposed to acquiring resistance to maraviroc (MVC) and other CCR5 antagonists in vitro has an inherent, low-level ability to utilize MVC-bound CCR5 for entry.Retrovirology. 2011 Nov 7;8:89. doi: 10.1186/1742-4690-8-89. Retrovirology. 2011. PMID: 22054077 Free PMC article.
-
HIV-1 entry and its inhibition.Curr Top Microbiol Immunol. 2003;281:1-27. doi: 10.1007/978-3-642-19012-4_1. Curr Top Microbiol Immunol. 2003. PMID: 12932074 Review.
-
[Viral entry as therapeutic target. Current situation of entry inhibitors].Enferm Infecc Microbiol Clin. 2008 Oct;26 Suppl 11:5-11. doi: 10.1016/s0213-005x(08)76557-1. Enferm Infecc Microbiol Clin. 2008. PMID: 19133215 Review. Spanish.
Cited by
-
A Protocol for Studying HIV-1 Envelope Glycoprotein Function.STAR Protoc. 2020 Oct 14;1(3):100133. doi: 10.1016/j.xpro.2020.100133. eCollection 2020 Dec 18. STAR Protoc. 2020. PMID: 33377027 Free PMC article.
-
A novel luciferase-based assay for quantifying coronavirus-induced syncytia.Sci Rep. 2025 May 20;15(1):17423. doi: 10.1038/s41598-025-02037-4. Sci Rep. 2025. PMID: 40394090 Free PMC article.
-
Enhancing anti-viral neutralization response to immunization with HIV-1 envelope glycoprotein immunogens.NPJ Vaccines. 2023 Nov 24;8(1):181. doi: 10.1038/s41541-023-00774-z. NPJ Vaccines. 2023. PMID: 37996435 Free PMC article.
-
Peptide Triazole Thiol Irreversibly Inactivates Metastable HIV-1 Env by Accessing Conformational Triggers Intrinsic to Virus-Cell Entry.Microorganisms. 2021 Jun 12;9(6):1286. doi: 10.3390/microorganisms9061286. Microorganisms. 2021. PMID: 34204725 Free PMC article.
-
Nanoluciferase-based cell fusion assay for rapid and high-throughput assessment of SARS-CoV-2-neutralizing antibodies in patient samples.Methods Enzymol. 2022;675:351-381. doi: 10.1016/bs.mie.2022.07.015. Epub 2022 Sep 9. Methods Enzymol. 2022. PMID: 36220277 Free PMC article.
References
-
- Barre-Sinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science. 1983;220:868–871. - PubMed
-
- Gallo RC, Salahuddin SZ, Popovic M, Shearer GM, Kaplan M, et al. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science. 1984;224:500–503. - PubMed
-
- Mehellou Y, De Clercq E. Twenty-six years of anti-HIV drug discovery: where do we stand and where do we go? J Med Chem. 2010;53:521–538. - PubMed
-
- Arenas-Pinto A, Grant A, Bhaskaran K, Copas A, Carr A, et al. Risk factors for fatality in HIV-infected patients with dideoxynucleoside-induced severe hyperlactataemia or lactic acidosis. Antivir Ther. 2011;16:219–226. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

