The molecular basis for the broad substrate specificity of human sulfotransferase 1A1
- PMID: 22069470
- PMCID: PMC3206062
- DOI: 10.1371/journal.pone.0026794
The molecular basis for the broad substrate specificity of human sulfotransferase 1A1
Abstract
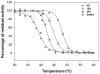
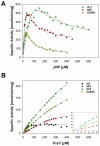
Cytosolic sulfotransferases (SULTs) are mammalian enzymes that detoxify a wide variety of chemicals through the addition of a sulfate group. Despite extensive research, the molecular basis for the broad specificity of SULTs is still not understood. Here, structural, protein engineering and kinetic approaches were employed to obtain deep understanding of the molecular basis for the broad specificity, catalytic activity and substrate inhibition of SULT1A1. We have determined five new structures of SULT1A1 in complex with different acceptors, and utilized a directed evolution approach to generate SULT1A1 mutants with enhanced thermostability and increased catalytic activity. We found that active site plasticity enables binding of different acceptors and identified dramatic structural changes in the SULT1A1 active site leading to the binding of a second acceptor molecule in a conserved yet non-productive manner. Our combined approach highlights the dominant role of SULT1A1 structural flexibility in controlling the specificity and activity of this enzyme.
Conflict of interest statement
Figures

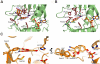


References
-
- Chapman E, Best MD, Hanson SR, Wong CH. Sulfotransferases: structure, mechanism, biological activity, inhibition, and synthetic utility. Angew Chem Int Ed Engl. 2004;43:3526–3548. - PubMed
-
- Gamage N, Barnett A, Hempel N, Duggleby RG, Windmill KF, et al. Human sulfotransferases and their role in chemical metabolism. Toxicol Sci. 2006;90:5–22. - PubMed
-
- Negishi M, Pedersen LG, Petrotchenko E, Shevtsov S, Gorokhov A, et al. Structure and function of sulfotransferases. Arch Biochem Biophys. 2001;390:149–157. - PubMed
-
- Wang Y, Spitz MR, Tsou AM, Zhang K, Makan N, et al. Sulfotransferase (SULT) 1A1 polymorphism as a predisposition factor for lung cancer: a case-control analysis. Lung Cancer. 2002;35:137–142. - PubMed
-
- Blanchard RL, Freimuth RR, Buck J, Weinshilboum RM, Coughtrie MW. A proposed nomenclature system for the cytosolic sulfotransferase (SULT) superfamily. Pharmacogenetics. 2004;14:199–211. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

