Biophysical analysis of apolipoprotein E3 variants linked with development of type III hyperlipoproteinemia
- PMID: 22069485
- PMCID: PMC3206067
- DOI: 10.1371/journal.pone.0027037
Biophysical analysis of apolipoprotein E3 variants linked with development of type III hyperlipoproteinemia
Abstract
Background: Apolipoprotein E (apoE) is a major protein of the lipoprotein transport system that plays important roles in lipid homeostasis and protection from atherosclerosis. ApoE is characterized by structural plasticity and thermodynamic instability and can undergo significant structural rearrangements as part of its biological function. Mutations in the 136-150 region of the N-terminal domain of apoE, reduce its low density lipoprotein (LDL) receptor binding capacity and have been linked with lipoprotein disorders, such as type III hyperlipoproteinemia (HLP) in humans. However, the LDL-receptor binding defects for these apoE variants do not correlate well with the severity of dyslipidemia, indicating that these variants may carry additional properties that contribute to their pathogenic potential.
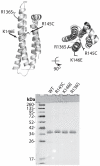
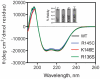
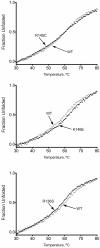
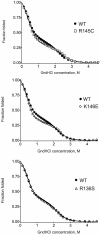
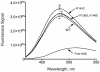
Methodology/principal findings: In this study we examined whether three type III HLP predisposing apoE3 variants, namely R136S, R145C and K146E affect the biophysical properties of the protein. Circular dichroism (CD) spectroscopy revealed that these mutations do not significantly alter the secondary structure of the protein. Thermal and chemical unfolding analysis revealed small thermodynamic alterations in each variant compared to wild-type apoE3, as well as effects in the reversibility of the unfolding transition. All variants were able to remodel multillamelar 1,2-Dimyristoyl-sn-glycero-3-phosphocholine (DMPC) vesicles, but R136S and R145C had reduced kinetics. Dynamic light scattering analysis indicated that the variant R136S exists in a higher-order oligomerization state in solution. Finally, 1-anilinonaphthalene-8-sulfonic acid (ANS) binding suggested that the variant R145C exposes a larger amount of hydrophobic surface to the solvent.
Conclusions/significance: Overall, our findings suggest that single amino acid changes in the functionally important region 136-150 of apoE3 can affect the molecule's stability and conformation in solution and may underlie functional consequences. However, the magnitude and the non-concerted nature of these changes, make it unlikely that they constitute a distinct unifying mechanism leading to type III HLP pathogenesis.
Conflict of interest statement
Figures



References
-
- Zannis VI, Kypreos KE, Chroni A, Kardassis D, Zanni EE. Abington, UK: Taylor & Francis; 2004. Lipoproteins and atherogenesis, in Molecular Mechanisms of Atherosclerosis J. L, editor.
-
- Kypreos KE, Zannis VI. LDL receptor deficiency or apoE mutations prevent remnant clearance and induce hypertriglyceridemia in mice. J Lipid Res. 2006;47:521–529. - PubMed
-
- Cooper AD. Hepatic uptake of chylomicron remnants. J Lipid Res. 1997;38:2173–2192. - PubMed
-
- Mahley RW, Ji ZS. Remnant lipoprotein metabolism: key pathways involving cell-surface heparan sulfate proteoglycans and apolipoprotein E. J Lipid Res. 1999;40:1–16. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

