The JAK-STAT pathway controls Plasmodium vivax load in early stages of Anopheles aquasalis infection
- PMID: 22069502
- PMCID: PMC3206008
- DOI: 10.1371/journal.pntd.0001317
The JAK-STAT pathway controls Plasmodium vivax load in early stages of Anopheles aquasalis infection
Abstract
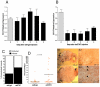
Malaria affects 300 million people worldwide every year and 450,000 in Brazil. In coastal areas of Brazil, the main malaria vector is Anopheles aquasalis, and Plasmodium vivax is responsible for the majority of malaria cases in the Americas. Insects possess a powerful immune system to combat infections. Three pathways control the insect immune response: Toll, IMD, and JAK-STAT. Here we analyze the immune role of the A. aquasalis JAK-STAT pathway after P. vivax infection. Three genes, the transcription factor Signal Transducers and Activators of Transcription (STAT), the regulatory Protein Inhibitors of Activated STAT (PIAS) and the Nitric Oxide Synthase enzyme (NOS) were characterized. Expression of STAT and PIAS was higher in males than females and in eggs and first instar larvae when compared to larvae and pupae. RNA levels for STAT and PIAS increased 24 and 36 hours (h) after P. vivax challenge. NOS transcription increased 36 h post infection (hpi) while this protein was already detected in some midgut epithelial cells 24 hpi. Imunocytochemistry experiments using specific antibodies showed that in non-infected insects STAT and PIAS were found mostly in the fat body, while in infected mosquitoes the proteins were found in other body tissues. The knockdown of STAT by RNAi increased the number of oocysts in the midgut of A. aquasalis. This is the first clear evidence for the involvement of a specific immune pathway in the interaction of the Brazilian malaria vector A. aquasalis with P. vivax, delineating a potential target for the future development of disease controlling strategies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



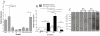




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

