Split-protein systems: beyond binary protein-protein interactions
- PMID: 22070901
- PMCID: PMC3237955
- DOI: 10.1016/j.cbpa.2011.10.014
Split-protein systems: beyond binary protein-protein interactions
Abstract
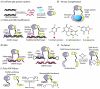
It has been estimated that 650,000 protein-protein interactions exist in the human interactome (Stumpf et al., 2008), a subset of all possible macromolecular partnerships that dictate life. Thus there is a continued need for the development of sensitive and user-friendly methods for cataloguing biomacromolecules in complex environments and for detecting their interactions, modifications, and cellular location. Such methods also allow for establishing differences in the interactome between a normal and diseased cellular state and for quantifying the outcome of therapeutic intervention. A promising approach for deconvoluting the role of macromolecular partnerships is split-protein reassembly, also called protein fragment complementation. This approach relies on the appropriate fragmentation of protein reporters, such as the green fluorescent protein or firefly luciferase, which when attached to possible interacting partners can reassemble and regain function, thereby confirming the partnership. Split-protein methods have been effectively utilized for detecting protein-protein interactions in cell-free systems, Escherichia coli, yeast, mammalian cells, plants, and live animals. Herein, we present recent advances in engineering split-protein systems that allow for the rapid detection of ternary protein complexes, small molecule inhibitors, as well as a variety of macromolecules including nucleic acids, poly(ADP) ribose, and iron sulfur clusters. We also present advances that combine split-protein systems with chemical inducers of dimerization strategies that allow for regulating the activity of orthogonal split-proteases as well as aid in identifying enzyme inhibitors. Finally, we discuss autoinhibition strategies leading to turn-on sensors as well as future directions in split-protein methodology including possible therapeutic approaches.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
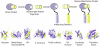

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

