Methanogenic archaea are globally ubiquitous in aerated soils and become active under wet anoxic conditions
- PMID: 22071343
- PMCID: PMC3309352
- DOI: 10.1038/ismej.2011.141
Methanogenic archaea are globally ubiquitous in aerated soils and become active under wet anoxic conditions
Abstract
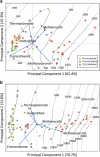
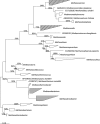
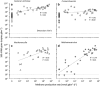
The prototypical representatives of the Euryarchaeota--the methanogens--are oxygen sensitive and are thought to occur only in highly reduced, anoxic environments. However, we found methanogens of the genera Methanosarcina and Methanocella to be present in many types of upland soils (including dryland soils) sampled globally. These methanogens could be readily activated by incubating the soils as slurry under anoxic conditions, as seen by rapid methane production within a few weeks, without any additional carbon source. Analysis of the archaeal 16S ribosomal RNA gene community profile in the incubated samples through terminal restriction fragment length polymorphism and quantification through quantitative PCR indicated dominance of Methanosarcina, whose gene copy numbers also correlated with methane production rates. Analysis of the δ(13)C of the methane further supported this, as the dominant methanogenic pathway was in most cases aceticlastic, which Methanocella cannot perform. Sequences of the key methanogenic enzyme methyl coenzyme M reductase retrieved from the soil samples before incubation confirmed that Methanosarcina and Methanocella are the dominant methanogens, though some sequences of Methanobrevibacter and Methanobacterium were also detected. The global occurrence of only two active methanogenic archaea supports the hypothesis that these are autochthonous members of the upland soil biome and are well adapted to their environment.
Figures

References
-
- Angel R, Conrad R. In situ measurement of methane fluxes and analysis of transcribed particulate methane monooxygenase in desert soils. Environ Microbiol. 2009;11:2598–2610. - PubMed
-
- Angel R, Soares MIM, Ungar ED, Gillor O. Biogeography of soil archaea and bacteria along a steep precipitation gradient. ISME J. 2010;4:553–563. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

