JC virus promoter/enhancers contain TATA box-associated Spi-B-binding sites that support early viral gene expression in primary astrocytes
- PMID: 22071512
- PMCID: PMC3352355
- DOI: 10.1099/vir.0.035832-0
JC virus promoter/enhancers contain TATA box-associated Spi-B-binding sites that support early viral gene expression in primary astrocytes
Abstract
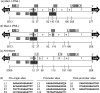
JC virus (JCV) is the aetiological agent of the demyelinating disease progressive multifocal leukoencephalopathy, an AIDS defining illness and serious complication of mAb therapies. Initial infection probably occurs in childhood. In the working model of dissemination, virus persists in the kidney and lymphoid tissues until immune suppression/modulation causes reactivation and trafficking to the brain where JCV replicates in oligodendrocytes. JCV infection is regulated through binding of host factors such as Spi-B to, and sequence variation in the non-coding control region (NCCR). Although NCCR sequences differ between sites of persistence and pathogenesis, evidence suggests that the virus that initiates infection in the brain disseminates via B-cells derived from latently infected haematopoietic precursors in the bone marrow. Spi-B binds adjacent to TATA boxes in the promoter/enhancer of the PML-associated JCV Mad-1 and Mad-4 viruses but not the non-pathogenic, kidney-associated archetype. The Spi-B-binding site of Mad-1/Mad-4 differs from that of archetype by a single nucleotide, AAAAGGGAAGGGA to AAAAGGGAAGGTA. Point mutation of the Mad-1 Spi-B site reduced early viral protein large T-antigen expression by up to fourfold. Strikingly, the reverse mutation in the archetype NCCR increased large T-antigen expression by 10-fold. Interestingly, Spi-B protein binds the NCCR sequence flanking the viral promoter/enhancer, but these sites are not essential for early viral gene expression. The effect of mutating Spi-B-binding sites within the JCV promoter/enhancer on early viral gene expression strongly suggests a role for Spi-B binding to the viral promoter/enhancer in the activation of early viral gene expression.
Figures

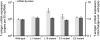


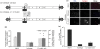
Similar articles
-
Transcription factor Spi-B binds unique sequences present in the tandem repeat promoter/enhancer of JC virus and supports viral activity.J Gen Virol. 2010 Dec;91(Pt 12):3042-52. doi: 10.1099/vir.0.023184-0. Epub 2010 Sep 8. J Gen Virol. 2010. PMID: 20826618 Free PMC article.
-
Propagation of archetype and nonarchetype JC virus variants in human fetal brain cultures: demonstration of interference activity by archetype JC virus.J Neurovirol. 2003 Oct;9(5):567-76. doi: 10.1080/13550280390241223. J Neurovirol. 2003. PMID: 13129771
-
Rearranged JC virus noncoding control regions found in progressive multifocal leukoencephalopathy patient samples increase virus early gene expression and replication rate.J Virol. 2010 Oct;84(20):10448-56. doi: 10.1128/JVI.00614-10. Epub 2010 Aug 4. J Virol. 2010. PMID: 20686041 Free PMC article.
-
The Role of the JC Virus in Central Nervous System Tumorigenesis.Int J Mol Sci. 2020 Aug 28;21(17):6236. doi: 10.3390/ijms21176236. Int J Mol Sci. 2020. PMID: 32872288 Free PMC article. Review.
-
Polyomavirus JC in the context of immunosuppression: a series of adaptive, DNA replication-driven recombination events in the development of progressive multifocal leukoencephalopathy.Clin Dev Immunol. 2013;2013:197807. doi: 10.1155/2013/197807. Epub 2013 Apr 15. Clin Dev Immunol. 2013. PMID: 23690820 Free PMC article. Review.
Cited by
-
Understanding Progressive Multifocal Leukoencephalopathy Risk in Multiple Sclerosis Patients Treated with Immunomodulatory Therapies: A Bird's Eye View.Front Immunol. 2018 Feb 2;9:138. doi: 10.3389/fimmu.2018.00138. eCollection 2018. Front Immunol. 2018. PMID: 29456537 Free PMC article. Review.
-
ViroFind: A novel target-enrichment deep-sequencing platform reveals a complex JC virus population in the brain of PML patients.PLoS One. 2018 Jan 23;13(1):e0186945. doi: 10.1371/journal.pone.0186945. eCollection 2018. PLoS One. 2018. PMID: 29360822 Free PMC article.
-
The Transcriptional Network Structure of a Myeloid Cell: A Computational Approach.Int J Genomics. 2017;2017:4858173. doi: 10.1155/2017/4858173. Epub 2017 Sep 30. Int J Genomics. 2017. PMID: 29119102 Free PMC article.
-
Imperfect Symmetry of Sp1 and Core Promoter Sequences Regulates Early and Late Virus Gene Expression of the Bidirectional BK Polyomavirus Noncoding Control Region.J Virol. 2016 Oct 28;90(22):10083-10101. doi: 10.1128/JVI.01008-16. Print 2016 Nov 15. J Virol. 2016. PMID: 27581987 Free PMC article.
-
New insights on human polyomavirus JC and pathogenesis of progressive multifocal leukoencephalopathy.Clin Dev Immunol. 2013;2013:839719. doi: 10.1155/2013/839719. Epub 2013 Apr 17. Clin Dev Immunol. 2013. PMID: 23690827 Free PMC article. Review.
References
-
- Amemiya K., Traub R., Durham L., Major E. O. (1989). Interaction of a nuclear factor-1-like protein with the regulatory region of the human polyomavirus JC virus. J Biol Chem 264, 7025–7032 - PubMed
-
- Amemiya K., Traub R., Durham L., Major E. O. (1992). Adjacent nuclear factor-1 and activator protein binding sites in the enhancer of the neurotropic JC virus. A common characteristic of many brain-specific genes. J Biol Chem 267, 14204–14211 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

