Identification of the first PAR1 deletion encompassing upstream SHOX enhancers in a family with idiopathic short stature
- PMID: 22071895
- PMCID: PMC3234524
- DOI: 10.1038/ejhg.2011.210
Identification of the first PAR1 deletion encompassing upstream SHOX enhancers in a family with idiopathic short stature
Abstract
Short stature homeobox-containing gene, MIM 312865 (SHOX) is located within the pseudoautosomal region 1 (PAR1) of the sex chromosomes. Mutations in SHOX or its downstream transcriptional regulatory elements represent the underlying molecular defect in ~60% of Léri-Weill dyschondrosteosis (LWD) and ~5-15% of idiopathic short stature (ISS) patients. Recently, three novel enhancer elements have been identified upstream of SHOX but to date, no PAR1 deletions upstream of SHOX have been observed that only encompass these enhancers in LWD or ISS patients. We set out to search for genetic alterations of the upstream SHOX regulatory elements in 63 LWD and 100 ISS patients with no known alteration in SHOX or the downstream enhancer regions using a specifically designed MLPA assay, which covers the PAR1 upstream of SHOX. An upstream SHOX deletion was identified in an ISS proband and her affected father. The deletion was confirmed and delimited by array-CGH, to extend ~286 kb. The deletion included two of the upstream SHOX enhancers without affecting SHOX. The 13.3-year-old proband had proportionate short stature with normal GH and IGF-I levels. In conclusion, we have identified the first PAR1 deletion encompassing only the upstream SHOX transcription regulatory elements in a family with ISS. The loss of these elements may result in SHOX haploinsufficiency because of decreased SHOX transcription. Therefore, this upstream region should be included in the routine analysis of PAR1 in patients with LWD, LMD and ISS.
Figures
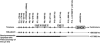

References
-
- Rao E, Weiss B, Fukami M, et al. Pseudoautosomal deletions encompassing a novel homeobox gene cause growth failure in idiopathic short stature and Turner syndrome. Nat Genet. 1997;16:54–63. - PubMed
-
- Ellison JW, Wardak Z, Young MF, Gehron Robey P, Laig-Webster M, Chiong W. PHOG, a candidate gene for involvement in the short stature of Turner syndrome. Hum Mol Genet. 1997;6:1341–1347. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials

