Sequence-based analysis uncovers an abundance of non-coding RNA in the total transcriptome of Mycobacterium tuberculosis
- PMID: 22072964
- PMCID: PMC3207917
- DOI: 10.1371/journal.ppat.1002342
Sequence-based analysis uncovers an abundance of non-coding RNA in the total transcriptome of Mycobacterium tuberculosis
Abstract
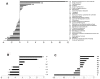
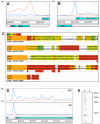
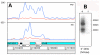
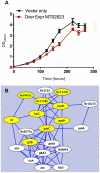
RNA sequencing provides a new perspective on the genome of Mycobacterium tuberculosis by revealing an extensive presence of non-coding RNA, including long 5' and 3' untranslated regions, antisense transcripts, and intergenic small RNA (sRNA) molecules. More than a quarter of all sequence reads mapping outside of ribosomal RNA genes represent non-coding RNA, and the density of reads mapping to intergenic regions was more than two-fold higher than that mapping to annotated coding sequences. Selected sRNAs were found at increased abundance in stationary phase cultures and accumulated to remarkably high levels in the lungs of chronically infected mice, indicating a potential contribution to pathogenesis. The ability of tubercle bacilli to adapt to changing environments within the host is critical to their ability to cause disease and to persist during drug treatment; it is likely that novel post-transcriptional regulatory networks will play an important role in these adaptive responses.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- WHO . Geneva: WHO; 2009. Global tuberculosis control - epidemiology, strategy, financing.
-
- Manganelli R, Voskuil MI Schoolnik GK, Dubnau E, Gomez M, et al. Role of the extracytoplasmic-function sigma factor sigma(H) in Mycobacterium tuberculosis global gene expression. Mol Microbiol. 2002;45:365–374. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

