Over-expression of DSCAM and COL6A2 cooperatively generates congenital heart defects
- PMID: 22072978
- PMCID: PMC3207880
- DOI: 10.1371/journal.pgen.1002344
Over-expression of DSCAM and COL6A2 cooperatively generates congenital heart defects
Abstract
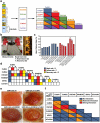
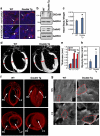
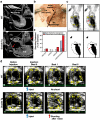
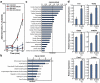
A significant current challenge in human genetics is the identification of interacting genetic loci mediating complex polygenic disorders. One of the best characterized polygenic diseases is Down syndrome (DS), which results from an extra copy of part or all of chromosome 21. A short interval near the distal tip of chromosome 21 contributes to congenital heart defects (CHD), and a variety of indirect genetic evidence suggests that multiple candidate genes in this region may contribute to this phenotype. We devised a tiered genetic approach to identify interacting CHD candidate genes. We first used the well vetted Drosophila heart as an assay to identify interacting CHD candidate genes by expressing them alone and in all possible pairwise combinations and testing for effects on rhythmicity or heart failure following stress. This comprehensive analysis identified DSCAM and COL6A2 as the most strongly interacting pair of genes. We then over-expressed these two genes alone or in combination in the mouse heart. While over-expression of either gene alone did not affect viability and had little or no effect on heart physiology or morphology, co-expression of the two genes resulted in ≈50% mortality and severe physiological and morphological defects, including atrial septal defects and cardiac hypertrophy. Cooperative interactions between DSCAM and COL6A2 were also observed in the H9C2 cardiac cell line and transcriptional analysis of this interaction points to genes involved in adhesion and cardiac hypertrophy. Our success in defining a cooperative interaction between DSCAM and COL6A2 suggests that the multi-tiered genetic approach we have taken involving human mapping data, comprehensive combinatorial screening in Drosophila, and validation in vivo in mice and in mammalian cells lines should be applicable to identifying specific loci mediating a broad variety of other polygenic disorders.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Schenone A, Mancardi GL. Molecular basis of inherited neuropathies. Curr Opin Neurol. 1999;12:603–616. - PubMed
-
- Warner LE, Garcia CA, Lupski JR. Hereditary peripheral neuropathies: clinical forms, genetics, and molecular mechanisms. Annu Rev Med. 1999;50:263–275. - PubMed
-
- Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010;61:437–455. - PubMed
-
- Couzin J. Genomics. The HapMap gold rush: researchers mine a rich deposit. Science. 2006;312:1131. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

