Multiple geographic origins of commensalism and complex dispersal history of Black Rats
- PMID: 22073158
- PMCID: PMC3206810
- DOI: 10.1371/journal.pone.0026357
Multiple geographic origins of commensalism and complex dispersal history of Black Rats
Abstract
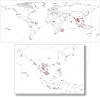
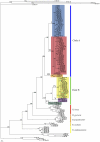
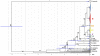
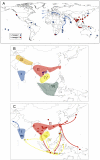
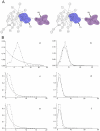
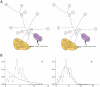
The Black Rat (Rattus rattus) spread out of Asia to become one of the world's worst agricultural and urban pests, and a reservoir or vector of numerous zoonotic diseases, including the devastating plague. Despite the global scale and inestimable cost of their impacts on both human livelihoods and natural ecosystems, little is known of the global genetic diversity of Black Rats, the timing and directions of their historical dispersals, and the risks associated with contemporary movements. We surveyed mitochondrial DNA of Black Rats collected across their global range as a first step towards obtaining an historical genetic perspective on this socioeconomically important group of rodents. We found a strong phylogeographic pattern with well-differentiated lineages of Black Rats native to South Asia, the Himalayan region, southern Indochina, and northern Indochina to East Asia, and a diversification that probably commenced in the early Middle Pleistocene. We also identified two other currently recognised species of Rattus as potential derivatives of a paraphyletic R. rattus. Three of the four phylogenetic lineage units within R. rattus show clear genetic signatures of major population expansion in prehistoric times, and the distribution of particular haplogroups mirrors archaeologically and historically documented patterns of human dispersal and trade. Commensalism clearly arose multiple times in R. rattus and in widely separated geographic regions, and this may account for apparent regionalism in their associated pathogens. Our findings represent an important step towards deeper understanding the complex and influential relationship that has developed between Black Rats and humans, and invite a thorough re-examination of host-pathogen associations among Black Rats.
Conflict of interest statement
Figures

References
-
- Barnett SA. The Story of Rats. Their impact on us, and our impact on them. 2001. 202 Allen & Unwin: Crows Nest, Sydney.
-
- Meerburg BG, Singleton GR, Leirs H. The year of the Rat ends—time to fight hunger! Pest Manag Sci. 2009;65:351–2. - PubMed
-
- Goodman SM. Rattus on Madagascar and the dilemma of protecting the endemic rodent fauna. Conserv Biol. 1994;9:450–3.
-
- Atkinson IAE. Moors PJ, editor. The spread of commensal species of Rattus to oceanic islands and their effects on island avifaunas. Conservation of island birds. 1985. pp. 35–81. International Council for Bird Preservation: Bristol.
-
- Duplantier J-M, Catalan J, Orth A, Grolleau B, Britton-Davidian J. Systematics of the black rat in Madagascar: consequences for the transmission and distribution of plague. Biol J Linn Soc. 2003;78:335–341.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

