Use of a high resolution melting (HRM) assay to compare gag, pol, and env diversity in adults with different stages of HIV infection
- PMID: 22073290
- PMCID: PMC3206918
- DOI: 10.1371/journal.pone.0027211
Use of a high resolution melting (HRM) assay to compare gag, pol, and env diversity in adults with different stages of HIV infection
Abstract
Background: Cross-sectional assessment of HIV incidence relies on laboratory methods to discriminate between recent and non-recent HIV infection. Because HIV diversifies over time in infected individuals, HIV diversity may serve as a biomarker for assessing HIV incidence. We used a high resolution melting (HRM) diversity assay to compare HIV diversity in adults with different stages of HIV infection. This assay provides a single numeric HRM score that reflects the level of genetic diversity of HIV in a sample from an infected individual.
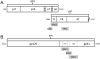
Methods: HIV diversity was measured in 203 adults: 20 with acute HIV infection (RNA positive, antibody negative), 116 with recent HIV infection (tested a median of 189 days after a previous negative HIV test, range 14-540 days), and 67 with non-recent HIV infection (HIV infected >2 years). HRM scores were generated for two regions in gag, one region in pol, and three regions in env.
Results: Median HRM scores were higher in non-recent infection than in recent infection for all six regions tested. In multivariate models, higher HRM scores in three of the six regions were independently associated with non-recent HIV infection.
Conclusions: The HRM diversity assay provides a simple, scalable method for measuring HIV diversity. HRM scores, which reflect the genetic diversity in a viral population, may be useful biomarkers for evaluation of HIV incidence, particularly if multiple regions of the HIV genome are examined.
Conflict of interest statement
Figures


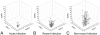

References
-
- Brookmeyer R. Measuring the HIV/AIDS epidemic: approaches and challenges. Epidemiol Rev. 2010;32:26–37. - PubMed
-
- Murphy G, Parry JV. Assays for the detection of recent infections with human immunodeficiency virus type 1. Euro Surveill. 2008;13 - PubMed
-
- Guy R, Gold J, Calleja JM, Kim AA, Parekh B, et al. Accuracy of serological assays for detection of recent infection with HIV and estimation of population incidence: a systematic review. Lancet Infect Dis. 2009;9:747–759. - PubMed
-
- Busch MP, Pilcher CD, Mastro TD, Kaldor J, Vercauteren G, et al. Beyond detuning: 10 years of progress and new challenges in the development and application of assays for HIV incidence estimation. AIDS. 2010;24:2763–2771. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AA016893/AA/NIAAA NIH HHS/United States
- N01 AI035173/AI/NIAID NIH HHS/United States
- U01 DA036935/DA/NIDA NIH HHS/United States
- R01 AI095068/AI/NIAID NIH HHS/United States
- R01-AI095068/AI/NIAID NIH HHS/United States
- R01-DA011602/DA/NIDA NIH HHS/United States
- U01-AI046745/AI/NIAID NIH HHS/United States
- R01-AA016893/AA/NIAAA NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- N01-AI35173/AI/NIAID NIH HHS/United States
- U01 AI068613/AI/NIAID NIH HHS/United States
- UM1 AI068613/AI/NIAID NIH HHS/United States
- N01-AI45200/AI/NIAID NIH HHS/United States
- U01-AI068613/AI/NIAID NIH HHS/United States
- R01 DA011602/DA/NIDA NIH HHS/United States
- N01-AI35176/AI/NIAID NIH HHS/United States
- N01 AI045200/AI/NIAID NIH HHS/United States
- UM1-AI068613/AI/NIAID NIH HHS/United States
- U01 AI046745/AI/NIAID NIH HHS/United States

